5F8W
 
 | | Crystal structure of a Crenomytilus grayanus lectin in complex with galactose | | Descriptor: | GLYCEROL, GalNAc/Gal-specific lectin, alpha-D-galactopyranose, ... | | Authors: | Liao, J.-H, Huang, K.-F, Tu, I.-F, Lee, I.-M, Wu, S.-H. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A Multivalent Marine Lectin from Crenomytilus grayanus Possesses Anti-cancer Activity through Recognizing Globotriose Gb3
J.Am.Chem.Soc., 138, 2016
|
|
2LAG
 
 | | Structure of the 44 kDa complex of interferon-alpha2 with the extracellular part of IFNAR2 obtained by 2D-double difference NOESY | | Descriptor: | Interferon alpha-2, Interferon alpha/beta receptor 2 | | Authors: | Nudelman, I, Akabayov, S.R, Scherf, T, Anglister, J. | | Deposit date: | 2011-03-13 | | Release date: | 2011-08-17 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Observation of Intermolecular Interactions in Large Protein Complexes by 2D-Double Difference Nuclear Overhauser Enhancement Spectroscopy: Application to the 44 kDa Interferon-Receptor Complex.
J.Am.Chem.Soc., 133, 2011
|
|
4C3J
 
 | | Structure of 14-subunit RNA polymerase I at 3.35 A resolution, crystal form C2-90 | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Fernandez-Tornero, C, Moreno-Morcillo, M, Rashid, U.J, Taylor, N.M.I, Ruiz, F.M, Gruene, T, Legrand, P, Steuerwald, U, Muller, C.W. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of the 14-Subunit RNA Polymerase I
Nature, 502, 2013
|
|
4V7F
 
 | | Arx1 pre-60S particle. | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
6T6K
 
 | | Y201W mutant of the orange carotenoid protein from Synechocystis at pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCINE, ... | | Authors: | Sluchanko, N.N, Gushchin, I, Botnarevskiy, V.S, Slonimskiy, Y.B, Remeeva, A, Kovalev, K, Stepanov, A.V, Gordeliy, V, Maksimov, E.G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Role of hydrogen bond alternation and charge transfer states in photoactivation of the Orange Carotenoid Protein.
Commun Biol, 4, 2021
|
|
5VFX
 
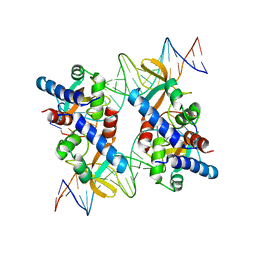 | | Structure of an accessory protein of the pCW3 relaxosome in complex with the origin of transfer (oriT) DNA | | Descriptor: | TcpK, oriT | | Authors: | Traore, D.A.K, Wisniewski, J.A, Flanigan, S.F, Conroy, P.J, Panjikar, S, Mok, Y.-F, Lao, C, Griffin, M.D.W, Adams, V, Rood, J.I, Whisstock, J.C. | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of TcpK in complex with oriT DNA of the antibiotic resistance plasmid pCW3.
Nat Commun, 9, 2018
|
|
1NM4
 
 | | Solution structure of Cu(I)-CopC from Pseudomonas syringae | | Descriptor: | Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Mangani, S, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-01-09 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A redox switch in CopC: An intriguing copper trafficking protein that binds copper(I) and copper(II)
at different sites
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5E8O
 
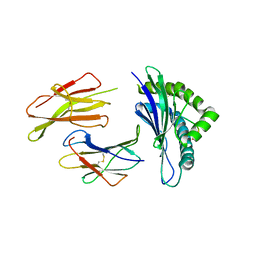 | | The structure of the TEIPP associated altered peptide ligand Trh4-p2ABU in complex with H-2D(b) | | Descriptor: | Beta-2-microglobulin, Ceramide synthase 5, H-2 class I histocompatibility antigen, ... | | Authors: | Hafstrand, I, Doorduijn, E, Duru, A.D, Buratto, J, Oliveira, C.C, Sandalova, T, van Hall, T, Achour, A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The MHC Class I Cancer-Associated Neoepitope Trh4 Linked with Impaired Peptide Processing Induces a Unique Noncanonical TCR Conformer.
J Immunol., 196, 2016
|
|
4C3H
 
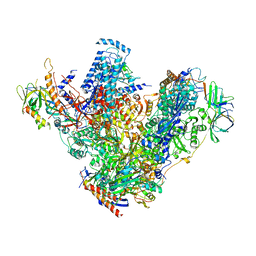 | | Structure of 14-subunit RNA polymerase I at 3.27 A resolution, crystal form C2-93 | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Fernandez-Tornero, C, Moreno-Morcillo, M, Rashid, U.J, Taylor, N.M.I, Ruiz, F.M, Gruene, T, Legrand, P, Steuerwald, U, Muller, C.W. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Crystal Structure of the 14-Subunit RNA Polymerase I
Nature, 502, 2013
|
|
1JLB
 
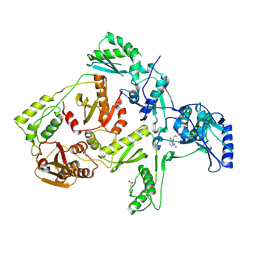 | | CRYSTAL STRUCTURE OF Y181C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
4C3I
 
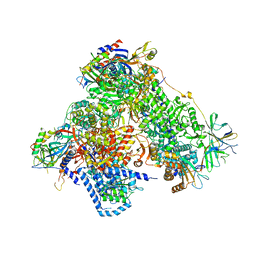 | | Structure of 14-subunit RNA polymerase I at 3.0 A resolution, crystal form C2-100 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, ... | | Authors: | Fernandez-Tornero, C, Moreno-Morcillo, M, Rashid, U.J, Taylor, N.M.I, Ruiz, F.M, Gruene, T, Legrand, P, Steuerwald, U, Muller, C.W. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the 14-Subunit RNA Polymerase I
Nature, 502, 2013
|
|
2N7L
 
 | |
1J1C
 
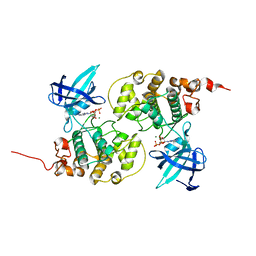 | | Binary complex structure of human tau protein kinase I with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase kinase-3 beta, MAGNESIUM ION | | Authors: | Aoki, M, Yokota, T, Sugiura, I, Sasaki, C, Hasegawa, T, Okumura, C, Kohno, T, Sugio, S, Matsuzaki, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into nucleotide recognition in tau-protein kinase I/glycogen synthase kinase 3 beta.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2CKB
 
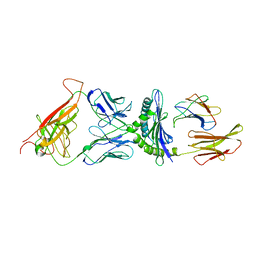 | | STRUCTURE OF THE 2C/KB/DEV8 COMPLEX | | Descriptor: | ALPHA, BETA T CELL RECEPTOR, BETA-2 MICROGLOBULIN, ... | | Authors: | Garcia, K.C, Degano, M, Wilson, I.A. | | Deposit date: | 1998-01-14 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen.
Science, 279, 1998
|
|
5VVR
 
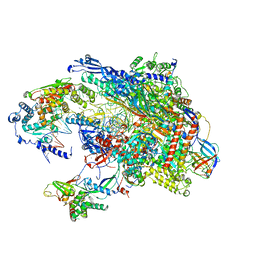 | |
1JLQ
 
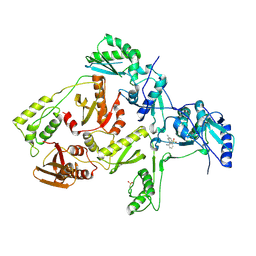 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 739W94 | | Descriptor: | 2-AMINO-6-(3,5-DIMETHYLPHENYL)SULFONYLBENZONITRILE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 2-Amino-6-arylsulfonylbenzonitriles as non-nucleoside reverse transcriptase inhibitors of HIV-1.
J.Med.Chem., 44, 2001
|
|
5E8P
 
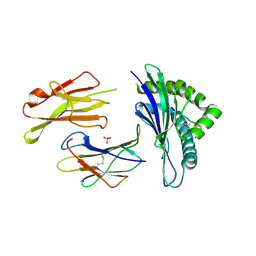 | | The structure of the TEIPP associated altered peptide ligand Trh4-p5NLE in complex with H-2D(b) | | Descriptor: | Beta-2-microglobulin, Ceramide synthase 5, H-2 class I histocompatibility antigen, ... | | Authors: | Hafstrand, I, Doorduijn, E, Duru, A.D, Buratto, J, Oliveira, C.C, Sandalova, T, van Hall, T, Achour, A. | | Deposit date: | 2015-10-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The MHC Class I Cancer-Associated Neoepitope Trh4 Linked with Impaired Peptide Processing Induces a Unique Noncanonical TCR Conformer.
J Immunol., 196, 2016
|
|
2KGF
 
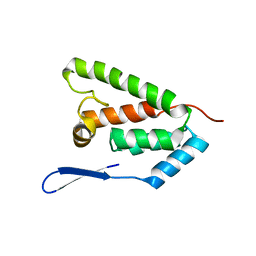 | | N-terminal domain of capsid protein from the Mason-Pfizer monkey virus | | Descriptor: | Capsid protein p27 | | Authors: | Macek, P, Chmelik, J, Zidek, L, Kaderavek, P, Padrta, P, Ruml, T, Pichova, I, Rumlova, M, Sklenar, V. | | Deposit date: | 2009-03-10 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the N-terminal domain of capsid protein from the mason-pfizer monkey virus
J.Mol.Biol., 392, 2009
|
|
1EZT
 
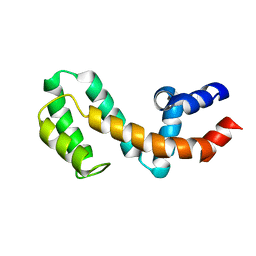 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 4 | | Authors: | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | Deposit date: | 2000-05-11 | | Release date: | 2001-01-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
3FON
 
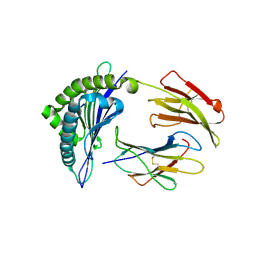 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide VNDIFEAI | | Descriptor: | Beta-2-microglobulin, MHC, Peptide | | Authors: | Malashkevich, V.N, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Brims, D.R, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect.
Int.Immunol., 22, 2010
|
|
1EZY
 
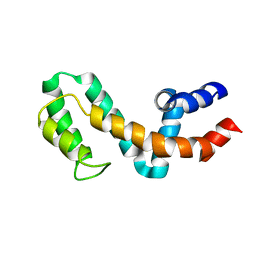 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 4 | | Authors: | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | Deposit date: | 2000-05-12 | | Release date: | 2001-01-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
2VBN
 
 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*AP*AP*AP*AP*GP*GP*CP*AP*GP*AP)-3', 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*AP*AP)-3', 5'-D(*TP*CP*TP*GP*CP*CP*TP*TP*TP*TP *TP*TP*GP*AP)-3', ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
2VBJ
 
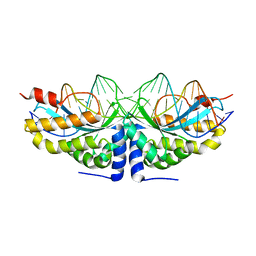 | | Molecular basis of human XPC gene recognition and cleavage by engineered homing endonuclease heterodimers | | Descriptor: | 5'-D(*TP*CP*TP*GP*CP*CP*TP*TP*TP*TP *TP*TP*GP*AP*AP*GP*GP*AP*TP*CP*CP*TP*AP*A)-3', 5'-D(*TP*TP*AP*GP*GP*AP*TP*CP*CP*TP *TP*CP*AP*AP*AP*AP*AP*AP*GP*GP*CP*AP*GP*A)-3', CALCIUM ION, ... | | Authors: | Redondo, P, Prieto, J, Munoz, I.G, Alibes, A, Stricher, F, Serrano, L, Arnould, S, Perez, C, Cabaniols, J.P, Duchateau, P, Paques, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2007-09-14 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis of Xeroderma Pigmentosum Group C DNA Recognition by Engineered Meganucleases
Nature, 456, 2008
|
|
2HRF
 
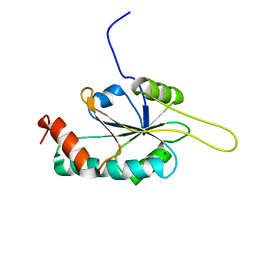 | | Solution Structure of Cu(I) P174L HSco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Leontari, I, Martinelli, M, Palumaa, P, Sillard, R, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human Sco1 functional studies and pathological implications of the P174L mutant.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HRN
 
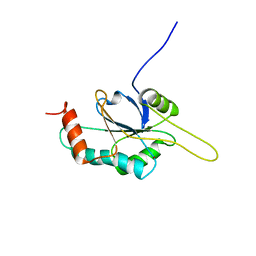 | | Solution Structure of Cu(I) P174L-HSco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Leontari, I, Martinelli, M, Palumaa, P, Sillard, R, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human Sco1 functional studies and pathological implications of the P174L mutant.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
