3VIF
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with gluconolactone | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-glucosidase, CHLORIDE ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SPX
 
 | | Crystal structure of O-Acetyl Serine Sulfhydrylase from Leishmania donovani | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, O-acetyl serine sulfhydrylase, ... | | Authors: | Raj, I, Gourinath, S. | | Deposit date: | 2011-07-04 | | Release date: | 2012-07-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The narrow active-site cleft of O-acetylserine sulfhydrylase from Leishmania donovani allows complex formation with serine acetyltransferases with a range of C-terminal sequences
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A1M
 
 | | NMR Structure of protoporphyrin-IX bound murine p22HBP | | Descriptor: | HEME-BINDING PROTEIN 1 | | Authors: | Goodfellow, B.J, Dias, J.S, Macedo, A.L, Ferreira, G.C, Peterson, F.C, Volkman, B.F, Duarte, I.C.N. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of Protoporphyrin-Ix Bound Murine P22Hbp
To be Published
|
|
3SVG
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | (1R)-1-[3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-ethoxyphenyl]ethanol, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hewings, S.D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Conway, S.J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-12 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand
TO BE PUBLISHED
|
|
3T97
 
 | |
3T7E
 
 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
4A67
 
 | | Mutations in the neighbourhood of CotA-laccase trinuclear site: D116E mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, SPORE COAT PROTEIN A | | Authors: | Silva, C.S, Lindley, P.F, Bento, I. | | Deposit date: | 2011-10-31 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Role of Asp116 in the Reductive Cleavage of Dioxygen to Water in Cota Laccase: Assistance During the Proton Transfer Mechanism
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4A9T
 
 | | CRYSTAL STRUCTURE OF HUMAN CHK2 IN COMPLEX WITH BENZIMIDAZOLE CARBOXAMIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-{[1-(4-CHLOROBENZYL)PIPERIDIN-4-YL]METHOXY}PHENYL)-1H-BENZIMIDAZOLE-5-CARBOXAMIDE, NITRATE ION, ... | | Authors: | Matijssen, C, Silva-Santisteban, M.C, Westwood, I.M, Siddique, S, Choi, V, Sheldrake, P, van Montfort, R.L.M, Blagg, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Benzimidazole Inhibitors of the Protein Kinase Chk2: Clarification of the Binding Mode by Flexible Side Chain Docking and Protein-Ligand Crystallography.
Bioorg.Med.Chem., 20, 2012
|
|
3T0G
 
 | | IspH:HMBPP (substrate) structure of the T167C mutant | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
3ZIG
 
 | | SepF-like protein from Pyrococcus furiosus | | Descriptor: | SEPF-LIKE PROTEIN | | Authors: | Duman, R, Ishikawa, S, Celik, I, Ogasawara, N, Lowe, J, Hamoen, L.W. | | Deposit date: | 2013-01-09 | | Release date: | 2013-11-27 | | Last modified: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Genetic Analyses Reveal the Protein Sepf as a New Membrane Anchor for the Z Ring.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3Q7B
 
 | | Exonuclease domain of Lassa virus nucleoprotein | | Descriptor: | Nucleoprotein, PHOSPHATE ION, ZINC ION | | Authors: | Hastie, K.M, Kimberlin, C.R, Zandonatti, M.A, MacRae, I.J, Saphire, E.O. | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3' to 5' exonuclease activity essential for immune suppression.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3X0R
 
 | | DP ribose pyrophosphatase from Thermus thermophilus HB8 in E'-state at reaction time of 30 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
3X0O
 
 | | ADP ribose pyrophosphatase from Thermus thermophilus HB8 in ESMM-state at reaction time of 10 min | | Descriptor: | ADP-ribose pyrophosphatase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Furuike, Y, Akita, Y, Miyahara, I, Kamiya, N. | | Deposit date: | 2014-10-17 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | ADP-Ribose Pyrophosphatase Reaction in Crystalline State Conducted by Consecutive Binding of Two Manganese(II) Ions as Cofactors
Biochemistry, 55, 2016
|
|
4A6W
 
 | | X-ray structures of oxazole hydroxamate EcMetAp-Mn complexes | | Descriptor: | 5-(2-chlorophenyl)-N-hydroxy-1,3-oxazole-2-carboxamide, MANGANESE (II) ION, METHIONINE AMINOPEPTIDASE | | Authors: | Huguet, F, Melet, A, AlvesdeSousa, R, Lieutaud, A, Chevalier, J, Deschamps, P, Tomas, A, Leulliot, N, Pages, J.M, Artaud, I. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Hydroxamic Acids as Potent Inhibitors of Fe(II) and Mn(II) E. Coli Methionine Aminopeptidase: Biological Activities and X-Ray Structures of Oxazole Hydroxamate-Ecmetap-Mn Complexes.
Chemmedchem, 7, 2012
|
|
3TWC
 
 | |
3TKM
 
 | | Crystal structure PPAR delta binding GW0742 | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor delta, {4-[({2-[3-fluoro-4-(trifluoromethyl)phenyl]-4-methyl-1,3-thiazol-5-yl}methyl)sulfanyl]-2-methylphenoxy}acetic acid | | Authors: | Trivella, D.B.B, Batista, F.H, Polikarpov, I. | | Deposit date: | 2011-08-27 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structural Insights into Human Peroxisome Proliferator Activated Receptor Delta (PPAR-Delta) Selective Ligand Binding.
Plos One, 7, 2012
|
|
3TMO
 
 | | The catalytic domain of human deubiquitinase DUBA | | Descriptor: | OTU domain-containing protein 5 | | Authors: | Yin, J, Bosanac, I, Ma, X, Hymowitz, S, Starovasnik, M, Cochran, A. | | Deposit date: | 2011-08-31 | | Release date: | 2012-01-11 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phosphorylation-dependent activity of the deubiquitinase DUBA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VF1
 
 | | Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca-regulation | | Descriptor: | 11R-lipoxygenase, FE (II) ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Eek, P, Jarving, R, Jarving, I, Gilbert, N.C, Newcomer, M.E, Samel, N. | | Deposit date: | 2012-01-09 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structure of a Calcium-dependent 11R-Lipoxygenase Suggests a Mechanism for Ca2+ Regulation.
J.Biol.Chem., 287, 2012
|
|
3VIO
 
 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with a new glucopyranosidic product | | Descriptor: | 3-{4-[2-(beta-D-glucopyranosyloxy)ethyl]piperazin-1-yl}propane-1-sulfonic acid, Beta-glucosidase, CHLORIDE ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VLM
 
 | | Crystal Structure Analysis of the Met244Ala Variant of KatG from Haloarcula marismortui | | Descriptor: | Catalase-peroxidase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sato, T, Ten-i, T, Higuchi, W, Yoshimatsu, K, Fujiwara, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure and Kinetic Studies on Met244Ala Variant of KatG from HALOARCULA MARISMORTUI
To be Published
|
|
3U5K
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with Midazolam | | Descriptor: | 8-chloro-6-(2-fluorophenyl)-1-methyl-4H-imidazo[1,5-a][1,4]benzodiazepine, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Benzodiazepines and benzotriazepines as protein interaction inhibitors targeting bromodomains of the BET family.
Bioorg.Med.Chem., 20, 2012
|
|
3RK4
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2011-04-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3VBU
 
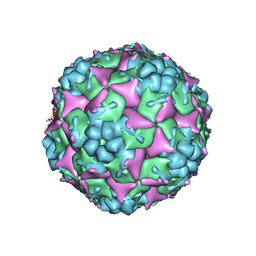 | | Crystal structure of empty human Enterovirus 71 particle | | Descriptor: | Genome Polyprotein, capsid protein VP0, capsid protein VP1, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VIL
 
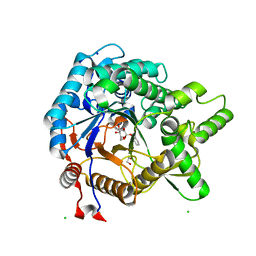 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with salicin | | Descriptor: | 1,2-ETHANEDIOL, 2-(hydroxymethyl)phenol, 2-(hydroxymethyl)phenyl beta-D-glucopyranoside, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VJA
 
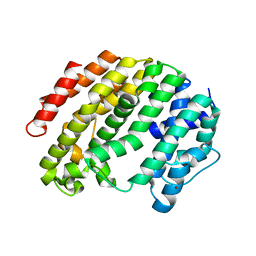 | | Crystal structure of the human squalene synthase | | Descriptor: | NICKEL (II) ION, Squalene synthase | | Authors: | Liu, C.I, Jeng, W.Y, Chang, W.J, Wang, A.H.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-04-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Binding modes of zaragozic acid A to human squalene synthase and staphylococcal dehydrosqualene synthase
J.Biol.Chem., 287, 2012
|
|
