5X12
 
 | | Crystal structure of Bacillus subtilis PadR | | Descriptor: | Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-01-24 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR
Nucleic Acids Res., 45, 2017
|
|
5X15
 
 | |
3C3Q
 
 | | ALIX Bro1-domain:CHMIP4B co-crystal structure | | Descriptor: | Charged multivesicular body protein 4b peptide, GLYCEROL, Programmed cell death 6-interacting protein | | Authors: | McCullough, J.B, Fisher, R.D, Whitby, F.G, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ALIX-CHMP4 interactions in the human ESCRT pathway.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1SRR
 
 | | CRYSTAL STRUCTURE OF A PHOSPHATASE RESISTANT MUTANT OF SPORULATION RESPONSE REGULATOR SPO0F FROM BACILLUS SUBTILIS | | Descriptor: | CALCIUM ION, SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Whiteley, J.M, Hoch, J.A, Zapf, J, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1996-04-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis.
Structure, 4, 1996
|
|
1LPC
 
 | |
2O3V
 
 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with paromamine derivative NB33 | | Descriptor: | (2S,3R,4R,5S,6R)-3-AMINO-4-({[(2S,3R,4R,5S,6R)-3-AMINO-2-{[(1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYL]OXY}-5-HYDROXY-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-4-YL]OXY}METHOXY)-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-2,5-DIOL, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
3RBH
 
 | | Structure of alginate export protein AlgE from Pseudomonas aeruginosa | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, Alginate production protein AlgE, ... | | Authors: | Whitney, J.C, Hay, I.D, Li, C, Eckford, P.D, Robinson, H, Amaya, M.F, Wood, L.F, Ohman, D.E, Bear, C.E, Rehm, B.H, Howell, P.L. | | Deposit date: | 2011-03-29 | | Release date: | 2011-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for alginate secretion across the bacterial outer membrane.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2FPK
 
 | | RadA recombinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Methanococcus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
1LP8
 
 | |
1SSD
 
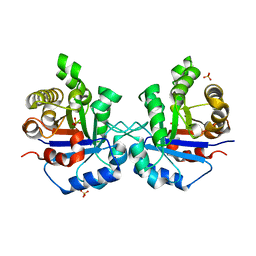 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
3I3G
 
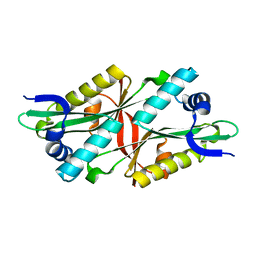 | | Crystal Structure of Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A | | Descriptor: | N-acetyltransferase | | Authors: | Qiu, W, Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, J Ferguson, M.A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A
To be Published
|
|
1SUR
 
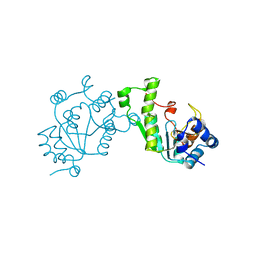 | | PHOSPHO-ADENYLYL-SULFATE REDUCTASE | | Descriptor: | PAPS REDUCTASE | | Authors: | Sinning, I, Savage, H. | | Deposit date: | 1998-04-01 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphoadenylyl sulphate (PAPS) reductase: a new family of adenine nucleotide alpha hydrolases.
Structure, 5, 1997
|
|
2FSJ
 
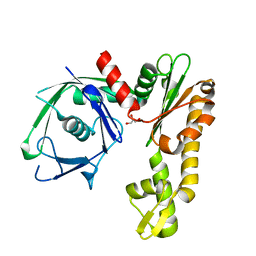 | | Crystal structure of Ta0583, an archaeal actin homolog, native data | | Descriptor: | GLYCEROL, hypothetical protein Ta0583 | | Authors: | Roeben, A, Kofler, C, Nagy, I, Nickell, S, Ulrich Hartl, F, Bracher, A. | | Deposit date: | 2006-01-23 | | Release date: | 2006-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal actin homolog
J.Mol.Biol., 358, 2006
|
|
5YU5
 
 | | Crystal structure of shaft pilin spaD from Lactobacillus rhamnosus GG | | Descriptor: | Pilus assembly protein | | Authors: | Chaurasia, P, Pratap, S, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2017-11-20 | | Release date: | 2018-06-20 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism.
Commun Biol, 1, 2018
|
|
3RQ8
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with P1,P5-Di(adenosine-5') pentaphosphate | | Descriptor: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RSQ
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, POTASSIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
5B09
 
 | | Polyketide cyclase OAC from Cannabis sativa bound with Olivetolic acid | | Descriptor: | 2,4-bis(oxidanyl)-6-pentyl-benzoic acid, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
2FO0
 
 | | Organization of the SH3-SH2 Unit in Active and Inactive Forms of the c-Abl Tyrosine Kinase | | Descriptor: | 6-(2,6-DICHLOROPHENYL)-2-{[3-(HYDROXYMETHYL)PHENYL]AMINO}-8-METHYLPYRIDO[2,3-D]PYRIMIDIN-7(8H)-ONE, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Seeliger, M, Davies, J.M, Weis, W.I, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2006-01-12 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase.
Mol.Cell, 21, 2006
|
|
1L1P
 
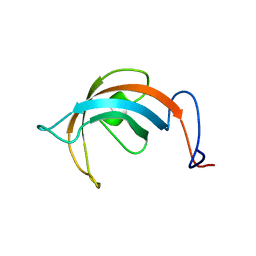 | | Solution Structure of the PPIase Domain from E. coli Trigger Factor | | Descriptor: | trigger factor | | Authors: | Kozlov, G, Trempe, J.-F, Perreault, A, Wong, M, Denisov, A, Ghandi, S, Gehring, K, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-02-19 | | Release date: | 2003-06-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Closed Form of a Peptidyl-Prolyl Isomerase Reveals the Mechanism of Protein Folding
To be Published
|
|
5B0G
 
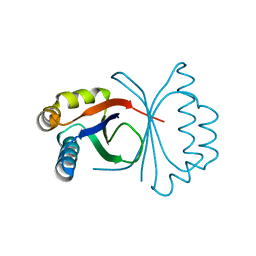 | | Polyketide cyclase OAC from Cannabis sativa, H78S mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
2FPM
 
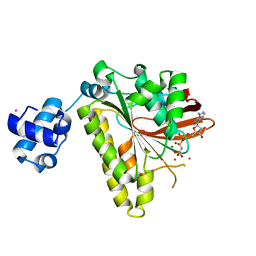 | | RadA recombinase in complex with AMP-PNP and high concentration of K+ | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2006-01-16 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Methanoccocus Voltae Rada in Complex with Adp: hydrolysis-induced conformational change
Biochemistry, 44, 2005
|
|
1T08
 
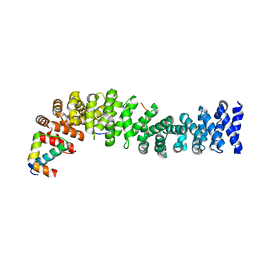 | | Crystal structure of beta-catenin/ICAT helical domain/unphosphorylated APC R3 | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin, Beta-catenin-interacting protein 1 | | Authors: | Ha, N.-C, Tonozuka, T, Stamos, J.L, Weis, W.I. | | Deposit date: | 2004-04-07 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation
Mol.Cell, 15, 2004
|
|
1L4M
 
 | | Crystal Structure of CobT complexed with 2-amino-p-cresol and nicotinate mononucleotide | | Descriptor: | 2-AMINO-P-CRESOL, NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica.
J.Biol.Chem., 277, 2002
|
|
2FRD
 
 | | Structure of Transhydrogenase (dI.S138A.NADH)2(dIII.NADPH)1 asymmetric complex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Brondijk, T.H, van Boxel, G.I, Mather, O.C, Quirk, P.G, White, S.A, Jackson, J.B. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Role of Invariant Amino Acid Residues at the Hydride Transfer Site of Proton-translocating Transhydrogenase.
J.Biol.Chem., 281, 2006
|
|
5Z43
 
 | | Crystal structure of prenyltransferase AmbP1 apo structure | | Descriptor: | AmbP1, MAGNESIUM ION | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
