3PIG
 
 | | beta-fructofuranosidase from Bifidobacterium longum | | Descriptor: | Beta-fructofuranosidase, CHLORIDE ION | | Authors: | Bujacz, A, Bujacz, G, Redzynia, I, Krzepkowska-Jedrzejczak, M, Bielecki, S. | | Deposit date: | 2010-11-06 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of the apo form of beta-fructofuranosidase from Bifidobacterium longum and its complex with fructose
Febs J., 278, 2011
|
|
5EXJ
 
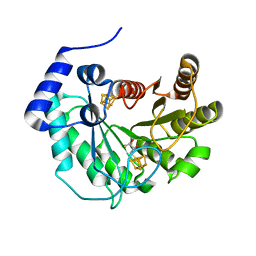 | | Crystal structure of M. tuberculosis lipoyl synthase at 1.64 A resolution | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Lipoyl synthase | | Authors: | McLaughlin, M.I, Lanz, N.D, Goldman, P.J, Lee, K.-H, Booker, S.J, Drennan, C.L. | | Deposit date: | 2015-11-23 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystallographic snapshots of sulfur insertion by lipoyl synthase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3VR3
 
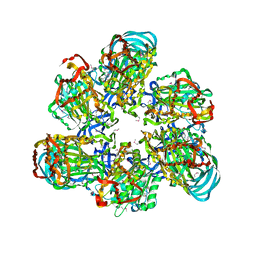 | | Crystal structure of AMP-PNP bound A3B3 complex from Enterococcus hirae V-ATPase [bA3B3] | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Arai, S, Saijo, S, Suzuki, K, Mizutani, K, Kakinuma, Y, Ishizuka-Katsura, Y, Ohsawa, N, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Yamato, I, Murata, T. | | Deposit date: | 2012-04-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Rotation mechanism of Enterococcus hirae V(1)-ATPase based on asymmetric crystal structures
Nature, 493, 2013
|
|
6ENZ
 
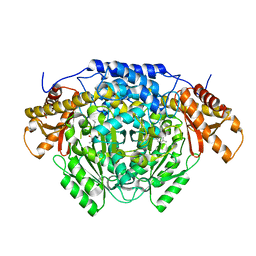 | | Crystal structure of mouse GADL1 | | Descriptor: | Acidic amino acid decarboxylase GADL1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Raasakka, A, Mahootchi, E, Winge, I, Luan, W, Kursula, P, Haavik, J. | | Deposit date: | 2017-10-07 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the mouse acidic amino acid decarboxylase GADL1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3VFB
 
 | | Crystal Structure of HIV-1 Protease Mutant N88D with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
1SSD
 
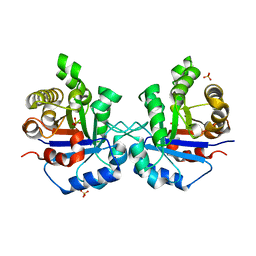 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
3VZN
 
 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35E mutant with Glu78 covalently bonded to 2-deoxy-2-fluoro-xylobiose | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
1SRR
 
 | | CRYSTAL STRUCTURE OF A PHOSPHATASE RESISTANT MUTANT OF SPORULATION RESPONSE REGULATOR SPO0F FROM BACILLUS SUBTILIS | | Descriptor: | CALCIUM ION, SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Whiteley, J.M, Hoch, J.A, Zapf, J, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1996-04-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis.
Structure, 4, 1996
|
|
3ESO
 
 | | Human transthyretin (TTR) complexed with N-(3,5-Dibromo-4-hydroxyphenyl)-2,5-dichlorobenzamide | | Descriptor: | 2,5-dichloro-N-(3,5-dibromo-4-hydroxyphenyl)benzamide, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2008-10-06 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Toward optimization of the second aryl substructure common to transthyretin amyloidogenesis inhibitors using biochemical and structural studies.
J.Med.Chem., 52, 2009
|
|
1SUR
 
 | | PHOSPHO-ADENYLYL-SULFATE REDUCTASE | | Descriptor: | PAPS REDUCTASE | | Authors: | Sinning, I, Savage, H. | | Deposit date: | 1998-04-01 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphoadenylyl sulphate (PAPS) reductase: a new family of adenine nucleotide alpha hydrolases.
Structure, 5, 1997
|
|
1T08
 
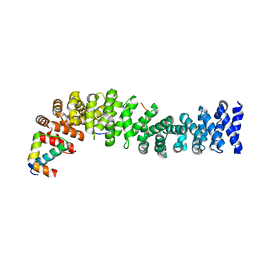 | | Crystal structure of beta-catenin/ICAT helical domain/unphosphorylated APC R3 | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin, Beta-catenin-interacting protein 1 | | Authors: | Ha, N.-C, Tonozuka, T, Stamos, J.L, Weis, W.I. | | Deposit date: | 2004-04-07 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation
Mol.Cell, 15, 2004
|
|
3QUJ
 
 | |
2MZ2
 
 | |
1TA6
 
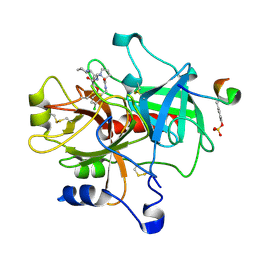 | | Crystal structure of thrombin in complex with compound 14b | | Descriptor: | 1-[2-AMINO-2-CYCLOHEXYL-ACETYL]-PYRROLIDINE-3-CARBOXYLIC ACID 5-CHLORO-2-(2-ETHYLCARBAMOYL-ETHOXY)-BENZYLAMIDE, Hirudin, thrombin | | Authors: | Tucker, T.J, Brady, S.F, Lumma, W.C, Lewis, S.D, Gardel, S.J, Naylor-Olsen, A.M, Yan, Y, Sisko, J.T, Stauffer, K.J, Lucas, B.Y, Lynch, J.J, Cook, J.J, Stranieri, M.T, Holahan, M.A, Lyle, E.A, Baskin, E.P, Chen, I.-W, Dancheck, K.B, Krueger, J.A, Cooper, C.M, Vacca, J.P. | | Deposit date: | 2004-05-19 | | Release date: | 2004-06-08 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of a series of potent and orally bioavailable
noncovalent thrombin inhibitors that utilize nonbasic groups in the P1 position
J.Med.Chem., 41, 1998
|
|
5F12
 
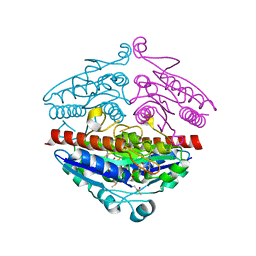 | | WrbA in complex with FMN under crystallization conditions of WrbA-FMN-BQ structure (4YQE) | | Descriptor: | FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone) | | Authors: | Degtjarik, O, Brynda, J, Ettrichova, O, Kuta Smatanova, I, Carey, J, Ettrich, R. | | Deposit date: | 2015-11-29 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Quantum Calculations Indicate Effective Electron Transfer between FMN and Benzoquinone in a New Crystal Structure of Escherichia coli WrbA.
J.Phys.Chem.B, 120, 2016
|
|
1T56
 
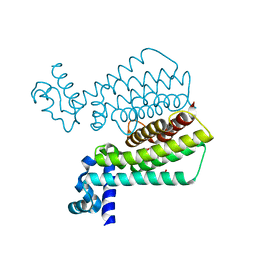 | | Crystal structure of TetR family repressor M. tuberculosis EthR | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, EthR repressor, GLYCEROL | | Authors: | Dover, L.G, Corsino, P.E, Daniels, I.R, Cocklin, S.L, Tatituri, V, Besra, G.S, Futterer, K. | | Deposit date: | 2004-05-03 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the TetR/CamR Family Repressor Mycobacterium tuberculosis EthR Implicated in Ethionamide Resistance.
J.Mol.Biol., 340, 2004
|
|
5EXI
 
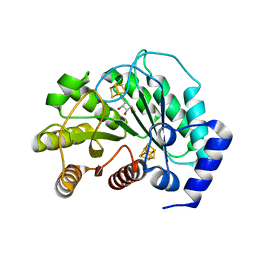 | | Crystal structure of M. tuberculosis lipoyl synthase at 2.28 A resolution | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Lipoyl synthase | | Authors: | McLaughlin, M.I, Lanz, N.D, Goldman, P.J, Lee, K.-H, Booker, S.J, Drennan, C.L. | | Deposit date: | 2015-11-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystallographic snapshots of sulfur insertion by lipoyl synthase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2N9Q
 
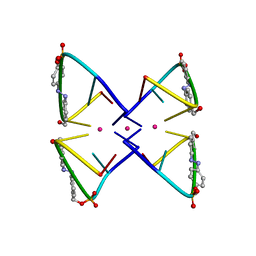 | | Photoswitchable G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*(AZW)P*GP*G)-3'), POTASSIUM ION | | Authors: | Thevarpadam, J, Bessi, I, Binas, O, Goncalves, D.P.N, Slavov, C, Jonker, H.R.A, Richter, C, Wachtveitl, J, Schwalbe, H, Heckel, A. | | Deposit date: | 2015-12-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Photoresponsive Formation of an Intermolecular Minimal G-Quadruplex Motif.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4Q58
 
 | |
2RMS
 
 | | Solution structure of the mSin3A PAH1-SAP25 SID complex | | Descriptor: | MSin3A-binding protein, Paired amphipathic helix protein Sin3a | | Authors: | Sahu, S.C, Swanson, K.A, Kang, R.S, Huang, K, Brubaker, K, Ratcliff, K, Radhakrishnan, I. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Conserved Themes in Target Recognition by the PAH1 and PAH2 Domains of the Sin3 Transcriptional Corepressor
J.Mol.Biol., 375, 2007
|
|
2RFN
 
 | | x-ray structure of c-Met with inhibitor. | | Descriptor: | 2-benzyl-5-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Kaplan-Lefko, P, Yang, Y, Zhang, Y, Moriguchi, J, Dussault, I. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | c-Met inhibitors with novel binding mode show activity against several hereditary papillary renal cell carcinoma-related mutations.
J.Biol.Chem., 283, 2008
|
|
2N1D
 
 | | Solution structure of the MRG15-MRGBP complex | | Descriptor: | MRG/MORF4L-binding protein, Mortality factor 4-like protein 1 | | Authors: | Xie, T, Zmysloski, A.M, Zhang, Y, Radhakrishnan, I. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Multi-specificity of MRG Domains.
Structure, 23, 2015
|
|
2N34
 
 | | NMR assignments and solution structure of the JAK interaction region of SOCS5 | | Descriptor: | Suppressor of cytokine signaling 5 | | Authors: | Chandrashekaran, I.R, Mohanty, B, Linossi, E.M, Nicholson, S.E, Babon, J, Norton, R.S, Dagley, L.F, Leung, E.W.W, Murphy, J.M. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Functional Characterization of the Conserved JAK Interaction Region in the Intrinsically Disordered N-Terminus of SOCS5.
Biochemistry, 54, 2015
|
|
2N4Y
 
 | | Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome | | Descriptor: | DNA_(5'-D(*CP*TP*GP*GP*GP*CP*GP*GP*GP*AP*CP*TP*GP*GP*GP*GP*AP*GP*TP*GP*GP*T)-3') | | Authors: | DeNicola, B, Lech, C.J, Heddi, B, Regmi, S, Frasson, I, Perrone, R, Richter, S.N, Phan, A.T. | | Deposit date: | 2015-07-02 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome.
Nucleic Acids Res., 44, 2016
|
|
3QWI
 
 | | Crystal structure of a 17beta-hydroxysteroid dehydrogenase (holo form) from fungus Cochliobolus lunatus in complex with NADPH and coumestrol | | Descriptor: | 1,2-ETHANEDIOL, 17beta-hydroxysteroid dehydrogenase, Coumestrol, ... | | Authors: | Cassetta, A, Lamba, D, Krastanova, I, Stojan, J, Lanisnik Rizner, T, Kristan, K, Brunskole, M. | | Deposit date: | 2011-02-28 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies on the flavonoid inhibition of a fungal 17Beta-Hydroxysteroid dehydrogenase
Biochem.J., 441, 2012
|
|
