3N6V
 
 | |
3RT9
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with Coenzyme A | | Descriptor: | COENZYME A, POTASSIUM ION, Putative uncharacterized protein, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
4Y1I
 
 | | Lactococcus lactis yybP-ykoY Mn riboswitch bound to Mn2+ | | Descriptor: | BARIUM ION, GUANOSINE-5'-TRIPHOSPHATE, Lactococcus lactis yybP-ykoY riboswitch, ... | | Authors: | Price, I.R, Ke, A. | | Deposit date: | 2015-02-07 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mn(2+)-Sensing Mechanisms of yybP-ykoY Orphan Riboswitches.
Mol.Cell, 57, 2015
|
|
4IVQ
 
 | | Crystal structure of thymidine kinase from Herpes simplex virus type 1 in complex with IN43/5 | | Descriptor: | 6-[3-hydroxy-2-(hydroxymethyl)prop-1-en-1-yl]-4-methoxy-1,5-dimethylpyrimidin-2(1H)-one, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Novakovic, I, Westermaier, Y, Perozzo, R, Raic-Malic, S, Scapozza, L. | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New thymine derivatives as HSV1-TK ligand for the development of PET imaging tracer
To be Published
|
|
4XQ3
 
 | | Crystal structure of Sso-SmAP2 | | Descriptor: | Like-Sm ribonucleoprotein core | | Authors: | Bezerra, G.A, Martens, B, Kreuter, M.J, Grishkovskaya, I, Manica, M, Arkhipova, V, Blasi, U, Djinovic-Carugo, K. | | Deposit date: | 2015-01-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Heptameric SmAP1 and SmAP2 Proteins of the Crenarchaeon Sulfolobus Solfataricus Bind to Common and Distinct RNA Targets.
Life, 5, 2015
|
|
2QGA
 
 | | Plasmodium vivax adenylosuccinate lyase Pv003765 with AMP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase, CALCIUM ION, ... | | Authors: | Lunin, V.V, Wernimont, A.K, Lew, J, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Sundstrom, M, Weigelt, J, Edwards, A.E, Hui, R, Hills, T, Altamentova, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Plasmodium vivax adenylosuccinate lyase Pv003765 with AMP bound
To be Published
|
|
3RFF
 
 | | Phosphopantetheine adenylyltransferase from Mycobacterium Tuberculosis (1.76 A resolution) | | Descriptor: | Phosphopantetheine adenylyltransferase | | Authors: | Timofeev, V.I, Smirnova, E.A, Chupova, L.A, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2011-04-06 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Three-Dimensional Structure of Phosphopantetheine Adenylyltransferase
from Mycobacterium Tuberculosis in the Apo Form and in Complexes
with Coenzyme A and D
Crystallography Reports, 57, 2012
|
|
3RRE
 
 | | Crystal Structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, GLYCEROL, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
4XXJ
 
 | | Crystal Structure of Escherichia coli-Expressed Halobacterium salinarum Bacteriorhodopsin in the Trimeric Form | | Descriptor: | Bacteriorhodopsin, EICOSANE, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Bratanov, D, Balandin, T, Round, E, Gushchin, I, Gordeliy, V. | | Deposit date: | 2015-01-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Approach to Heterologous Expression of Membrane Proteins. The Case of Bacteriorhodopsin.
Plos One, 10, 2015
|
|
4D6Z
 
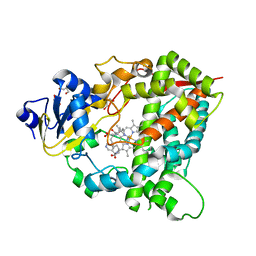 | |
4H8A
 
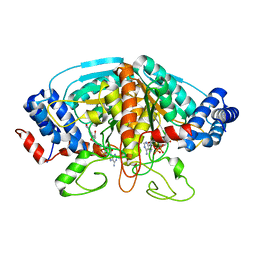 | | Crystal structure of ureidoglycolate dehydrogenase in binary complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Ureidoglycolate dehydrogenase | | Authors: | Rhee, S, Shin, I, Kim, M. | | Deposit date: | 2012-09-22 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural and functional insights into (s)-ureidoglycolate dehydrogenase, a metabolic branch point enzyme in nitrogen utilization.
Plos One, 7, 2012
|
|
3TEO
 
 | | APO Form of carbon disulfide hydrolase (selenomethionine form) | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, Carbon disulfide hydrolase | | Authors: | Smeulders, M.J, Barends, T.R.M.B, Pol, A, Scherer, A, Zandvoort, M.H, Udvarhelyi, A, Khadem, A, Menzel, A, Hermans, J, Shoeman, R.L, Wessels, H.J.C.T, van den Heuvel, L.P, Russ, L, Schlichting, I, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evolution of a new enzyme for carbon disulphide conversion by an acidothermophilic archaeon.
Nature, 478, 2011
|
|
4Y1J
 
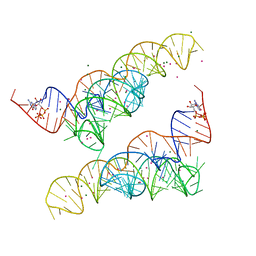 | |
2WHZ
 
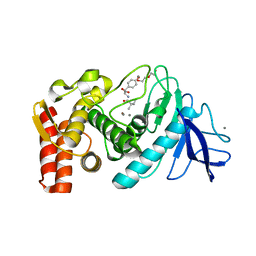 | |
4Y06
 
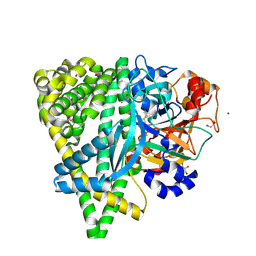 | | Crystal structure of the DAP BII (G675R) dipeptide complex | | Descriptor: | Dipeptidyl aminopeptidase BII, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Sakamoto, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
3RRF
 
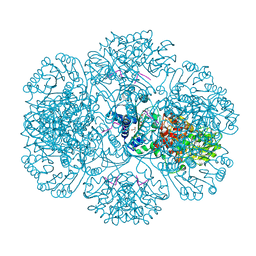 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bifunctional NAD(P)H-hydrate repair enzyme Nnr, GLYCEROL, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-04-29 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
4H2W
 
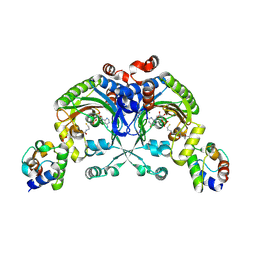 | | Crystal structure of engineered Bradyrhizobium japonicum glycine:[carrier protein] ligase complexed with carrier protein from Agrobacterium tumefaciens and AMP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, Amino acid--[acyl-carrier-protein] ligase 1, ... | | Authors: | Luic, M, Weygand-Durasevic, I, Ivic, N, Mocibob, M. | | Deposit date: | 2012-09-13 | | Release date: | 2013-03-06 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Adaptation of aminoacyl-tRNA synthetase catalytic core to carrier protein aminoacylation.
Structure, 21, 2013
|
|
3KUD
 
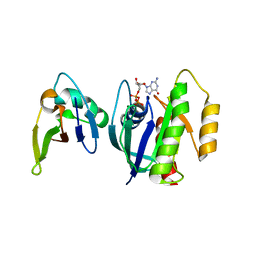 | | Complex of Ras-GDP with RafRBD(A85K) | | Descriptor: | GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Filchtinski, D, Sharabi, O, Rueppel, A, Vetter, I.R, Herrmann, C, Shifman, J.M. | | Deposit date: | 2009-11-27 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | What makes Ras an efficient molecular switch: a computational, biophysical, and structural study of Ras-GDP interactions with mutants of Raf.
J.Mol.Biol., 399, 2010
|
|
4H7M
 
 | |
5B4C
 
 | | Crystal structure of H10N mutant of LpxH with manganese | | Descriptor: | GLYCEROL, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Okada, C, Wakabayashi, H, Yao, M, Tanaka, I. | | Deposit date: | 2016-04-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structures of the UDP-diacylglucosamine pyrophosphohydrase LpxH from Pseudomonas aeruginosa
Sci Rep, 6, 2016
|
|
7ZG8
 
 | |
3ZL4
 
 | | Antibody structural organization: Role of kappa - lambda chain constant domain switch in catalytic functionality | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, A17 ANTIBODY FAB FRAGMENT HEAVY CHAIN, A17 ANTIBODY FAB FRAGMENT LAMBDA LIGHT CHAIN | | Authors: | Chatziefthimiou, S.D, Ponomarenko, N.A, Kurkova, I.N, Smirnov, A.V, Smirnov, I.V, Lamzin, V.S, Gabibov, A.G, Wilmanns, M. | | Deposit date: | 2013-01-28 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Role of Kappa>Lambda Light-Chain Constant-Domain Switch in the Structure and Functionality of A17 Reactibody
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3B9Q
 
 | | The crystal structure of cpFtsY from Arabidopsis thaliana | | Descriptor: | Chloroplast SRP receptor homolog, alpha subunit CPFTSY, MALONATE ION | | Authors: | Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2007-11-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of the chloroplast signal recognition particle (SRP) receptor reveals mechanistic details of SRP GTPase activation and a conserved membrane targeting site
Febs Lett., 581, 2007
|
|
3ZN6
 
 | | VP16-VP17 complex, a complex of the two major capsid proteins of bacteriophage P23-77 | | Descriptor: | CHLORIDE ION, SODIUM ION, VP16, ... | | Authors: | Rissanen, I, Grimes, J.M, Pawlowski, A, Mantynen, S, Harlos, K, Bamford, J.K.H, Stuart, D.I. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Structure, 21, 2013
|
|
3NAI
 
 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | 5-FLUOROURACIL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, K.H, Lee, J.H, Alam, I, Park, Y, Kang, S. | | Deposit date: | 2010-06-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase
To be Published
|
|
