3NH4
 
 | | Crystal structure of murine aminoacylase 3 | | Descriptor: | ACETATE ION, Aspartoacylase-2, CESIUM ION, ... | | Authors: | Hsieh, J.M, Tsirulnikov, K, Sawaya, M.R, Magilnick, N, Abuladze, N, Kurtz, I, Abramson, J, Pushkin, A. | | Deposit date: | 2010-06-14 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of aminoacylase 3 in complex with acetylated substrates.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4CDQ
 
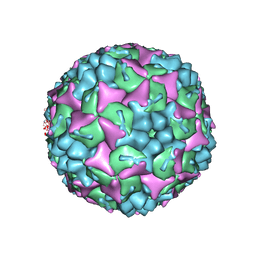 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP2 | | Descriptor: | 4-((5-(2-oxo-3-(pyridin-4-yl)imidazolidin-1-yl)pentyl)oxy)benzaldehyde O-ethyl oxime, SODIUM ION, VP1, ... | | Authors: | DeColibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4LHC
 
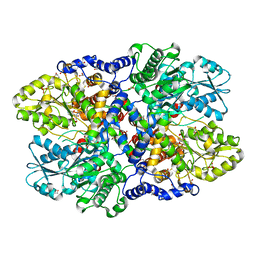 | | Crystal structure of Synechocystis sp. PCC 6803 glycine decarboxylase (P-protein), holo form with pyridoxal-5'-phosphate and glycine | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, BICINE, ... | | Authors: | Hasse, D, Andersson, E, Carlsson, G, Masloboy, A, Hagemann, M, Bauwe, H, Andersson, I. | | Deposit date: | 2013-07-01 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure of the Homodimeric Glycine Decarboxylase P-protein from Synechocystis sp. PCC 6803 Suggests a Mechanism for Redox Regulation.
J.Biol.Chem., 288, 2013
|
|
4LIS
 
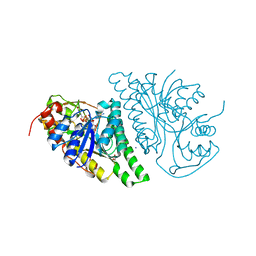 | | Crystal Structure of UDP-galactose-4-epimerase from Aspergillus nidulans | | Descriptor: | GLYCEROL, IODIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dalrymple, S.A, Ko, J, Sheoran, I, Kaminskyj, S.G.W, Sanders, D.A.R. | | Deposit date: | 2013-07-03 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Elucidation of Substrate Specificity in Aspergillus nidulans UDP-Galactose-4-Epimerase.
Plos One, 8, 2013
|
|
7ZFD
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-25 Fab | | Descriptor: | Omi-25 heavy chain, Omi-25 light chain, Spike protein S1 | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
3MES
 
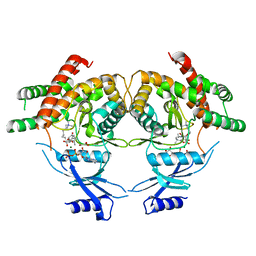 | | Crystal structure of choline kinase from Cryptosporidium parvum Iowa II, cgd3_2030 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Choline kinase, DECAMETHONIUM ION, ... | | Authors: | Qiu, W, Wernimont, A, Hills, T, Lew, J, Artz, J.D, Xiao, T, Allali-Hassani, A, Vedadi, M, Kozieradzki, I, Cossar, D, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Hui, R, Ma, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of choline kinase from Cryptosporidium parvum Iowa II, cgd3_2030
TO BE PUBLISHED
|
|
7ZFC
 
 | | SARS-CoV-2 Beta RBD in complex with nanobody C1, Omi-18 and Omi-31 Fabs | | Descriptor: | Nanobody C1, Omi-18 heavy chain, Omi-18 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF8
 
 | | SARS-CoV-2 Omicron BA.2 RBD in complex with COVOX-150 Fab | | Descriptor: | COVOX-150 heavy chain, COVOX-150 light chain, Spike protein S1 | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF9
 
 | | SARS-CoV-2 Omicron BA.2 RBD in complex with COVOX-150 Fab (P21) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-150 heavy chain, COVOX-150 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
5EZF
 
 | | Racemic crystal structures of Pribnow box consensus promoter sequence (Pbca) | | Descriptor: | CALCIUM ION, Complementary strand, Pribnow box template strand | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-11-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
7ZF4
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-9 Fab and nanobody F2 | | Descriptor: | Nanobody F2, Omi-9 heavy chain, Omi-9 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
4XC1
 
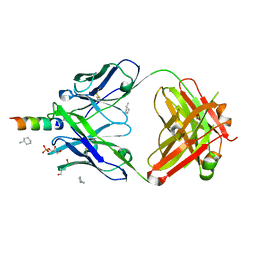 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 GP41: crystals cryoprotected with sn-Glycerol 3-phosphate | | Descriptor: | 4E10 FAB HEAVY CHAIN, 4E10 FAB LIGHT CHAIN, CYCLOHEXYLAMMONIUM ION, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
3MNV
 
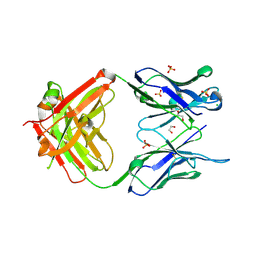 | | Crystal structure of the non-neutralizing HIV antibody 13H11 Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, ANTI-HIV-1 ANTIBODY 13H11 HEAVY CHAIN, ANTI-HIV-1 ANTIBODY 13H11 LIGHT CHAIN, ... | | Authors: | Nicely, N.I, Dennison, S.M, Kelsoe, G, Liao, H.-X, Alam, S.M, Haynes, B.F. | | Deposit date: | 2010-04-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a non-neutralizing antibody to the HIV-1 gp41 membrane-proximal external region.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7EFY
 
 | |
1BVY
 
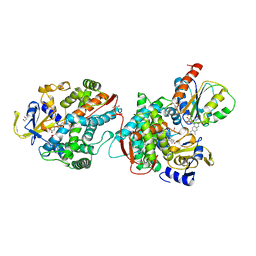 | | COMPLEX OF THE HEME AND FMN-BINDING DOMAINS OF THE CYTOCHROME P450(BM-3) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, PROTEIN (CYTOCHROME P450 BM-3), ... | | Authors: | Sevrioukova, I.F, Li, H, Zhang, H, Peterson, J.A, Poulos, T.L. | | Deposit date: | 1998-09-21 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of a cytochrome P450-redox partner electron-transfer complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4K9X
 
 | | Complex of human CYP3A4 with a desoxyritonavir analog | | Descriptor: | 1,3-thiazol-5-ylmethyl [(2R,5R)-5-{[(2S)-2-methylbutanoyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2013-04-21 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Dissecting Cytochrome P450 3A4-Ligand Interactions Using Ritonavir Analogues.
Biochemistry, 52, 2013
|
|
4XCE
 
 | |
3AFE
 
 | | Crystal structure of the HsaA monooxygenase from M.tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Eltis, L.D, Strynadka, N. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
1C4E
 
 | |
3M3D
 
 | | Crystal structure of Acetylcholinesterase in complex with Xenon | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Behnen, J, Brumshtein, B, Toker, L, Silman, I, Sussman, J.L, Klebe, G, Heine, A. | | Deposit date: | 2010-03-09 | | Release date: | 2011-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Old acquantance rediscover; use of xenon/protein complexes as a generic tool for SAD phasing of inhouse data
To be Published
|
|
7AZ7
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 37 bound | | Descriptor: | Beta sliding clamp, FORMIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
3MAV
 
 | | Crystal structure of Plasmodium vivax putative farnesyl pyrophosphate synthase (Pv092040) | | Descriptor: | Farnesyl pyrophosphate synthase, SULFATE ION | | Authors: | Dong, A, Dunford, J, Lew, J, Wernimont, A.K, Ren, H, Zhao, Y, Koeieradzki, I, Opperman, U, Sundstrom, M, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular characterization of a novel geranylgeranyl pyrophosphate synthase from Plasmodium parasites.
J.Biol.Chem., 286, 2011
|
|
3N2B
 
 | | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae. | | Descriptor: | CHLORIDE ION, Diaminopimelate decarboxylase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae.
To be Published
|
|
5F26
 
 | | Crystal structures of Pribnow box consensus promoter sequence (P63) | | Descriptor: | Complementary strand, Pribnow box consensus sequence strand | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Srivastava, S.C, Huc, I. | | Deposit date: | 2015-12-01 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure elucidation of the Pribnow box consensus promoter sequence by racemic DNA crystallography.
Nucleic Acids Res., 44, 2016
|
|
4HDR
 
 | | Crystal Structure of ArsAB in Complex with 5,6-dimethylbenzimidazole | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIMETHYLBENZIMIDAZOLE, ArsA, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
