6ZA1
 
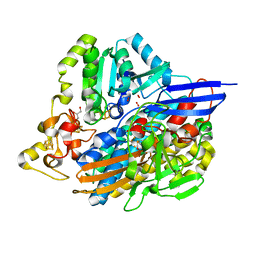 | | Structure of [NiFeSe] hydrogenase G491A variant from Desulfovibrio vulgaris Hildenborough pressurized with Oxygen gas - structure G491A-O2-hd | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, GLYCEROL, ... | | Authors: | Zacarias, S, Temporao, A, Carpentier, P, van der Linden, P, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Exploring the gas access routes in a [NiFeSe] hydrogenase using crystals pressurized with krypton and oxygen.
J.Biol.Inorg.Chem., 25, 2020
|
|
6ZI7
 
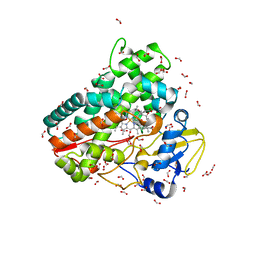 | | Crystal structure of OleP-oleandolide(DEO) bound to L-rhamnose | | Descriptor: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, Cytochrome P-450, FORMIC ACID, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | Deposit date: | 2020-06-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6YZZ
 
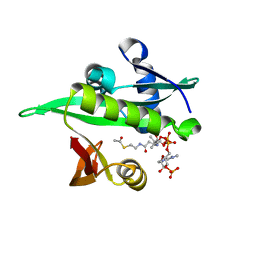 | | Arabidopsis thaliana Naa50 in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
6LX4
 
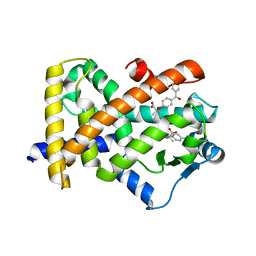 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX9
 
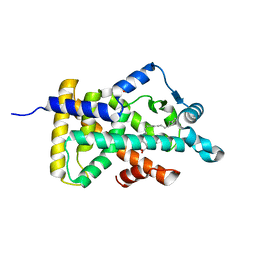 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
9OBP
 
 | | Cryo-EM structure of alpha-synuclein filaments derived from the frontal cortex of the case of atypical multiple system atrophy | | Descriptor: | Alpha-synuclein, Unidentified protein | | Authors: | Enomoto, M, Martinez-Valbuena, I, Forrest, S.L, Xu, X, Munhoz, R, Li, J, Rogaeva, E, Lang, A.E, Kovacs, G.G. | | Deposit date: | 2025-04-23 | | Release date: | 2025-06-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Lewy-MSA hybrid fold drives distinct neuronal alpha-synuclein pathology.
Commun Biol, 8, 2025
|
|
4U55
 
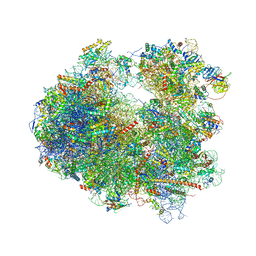 | | Crystal structure of Cryptopleurine bound to the yeast 80S ribosome | | Descriptor: | (14aR)-2,3,6-trimethoxy-11,12,13,14,14a,15-hexahydro-9H-dibenzo[f,h]pyrido[1,2-b]isoquinoline, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
9LSA
 
 | | Crystal structure of mRFP1 with a grafted calcium-binding sequence and one bound calcium ion in a calcium-containing solution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Red fluorescent protein,grafted calcium-binding sequence, ... | | Authors: | Uehara, R, Kamiya, Y, Maeda, S, Okamoto, K, Toya, S, Chiba, R, Amesaka, H, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Enhanced secretion through type 1 secretion system by grafting a calcium-binding sequence to modify the folding of cargo proteins.
Protein Sci., 34, 2025
|
|
9LSF
 
 | | Crystal structure of mRFP1 with a grafted calcium-binding sequence and one bound calcium ion in a calcium-free solution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uehara, R, Kamiya, Y, Maeda, S, Okamoto, K, Toya, S, Chiba, R, Amesaka, H, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2025-02-04 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Enhanced secretion through type 1 secretion system by grafting a calcium-binding sequence to modify the folding of cargo proteins.
Protein Sci., 34, 2025
|
|
9LSW
 
 | | Crystal structure of the calcium-free mRFP1 with a grafted calcium-binding sequence | | Descriptor: | Red fluorescent protein,grafted calcium-binding sequence | | Authors: | Uehara, R, Kamiya, Y, Maeda, S, Okamoto, K, Toya, S, Chiba, R, Amesaka, H, Takano, K, Matsumura, H, Tanaka, S.-i. | | Deposit date: | 2025-02-05 | | Release date: | 2025-06-04 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Enhanced secretion through type 1 secretion system by grafting a calcium-binding sequence to modify the folding of cargo proteins.
Protein Sci., 34, 2025
|
|
4U3N
 
 | | Crystal structure of CCA trinucleotide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U4G
 
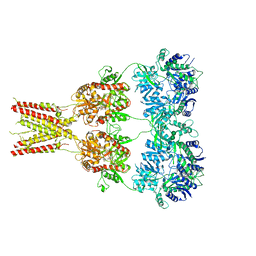 | | Structure of GluA2* in complex with competitive antagonist ZK 200775 | | Descriptor: | Glutamate receptor 2, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Yelshanskaya, M.V, Li, M, Sobolevsky, A.I. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.49 Å) | | Cite: | Structure of an agonist-bound ionotropic glutamate receptor.
Science, 345, 2014
|
|
4U4Q
 
 | | Crystal structure of Homoharringtonine bound to the yeast 80S ribosome | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
9PAP
 
 | |
4U8W
 
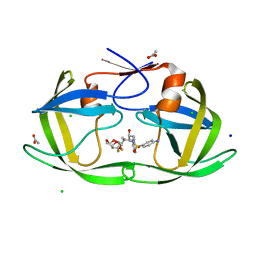 | | HIV-1 wild Type protease with GRL-050-10A (a Gem-difluoro-bis-Tetrahydrofuran as P2-Ligand) | | Descriptor: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of gem-Difluoro-bis-Tetrahydrofuran as P2 Ligand for HIV-1 Protease Inhibitors to Improve Brain Penetration: Synthesis, X-ray Studies, and Biological Evaluation.
Chemmedchem, 10, 2015
|
|
4U6F
 
 | | Crystal structure of T-2 toxin bound to the yeast 80S ribosome | | Descriptor: | 12,13-Epoxytrichothec-9-ene-3,4,8,15-tetrol-4,15-diacetate-8-isovalerate, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-28 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U4Z
 
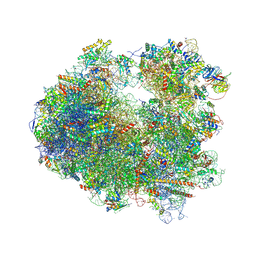 | | Crystal structure of Phyllanthoside bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
6T46
 
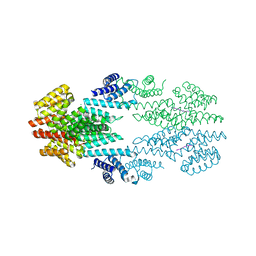 | | Structure of the Rap conjugation gene regulator of the plasmid pLS20 in complex with the Phr* peptide | | Descriptor: | CHLORIDE ION, Quorum-sensing secretion protein (processed), Response regulator aspartate phosphatase, ... | | Authors: | Crespo, I, Bernardo, N, Meijer, W.J.J, Boer, D.R. | | Deposit date: | 2019-10-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inactivation of the dimeric RappLS20 anti-repressor of the conjugation operon is mediated by peptide-induced tetramerization.
Nucleic Acids Res., 48, 2020
|
|
4U3U
 
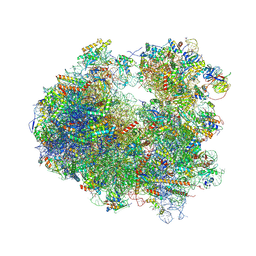 | | Crystal structure of Cycloheximide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U4U
 
 | | Crystal structure of Lycorine bound to the yeast 80S ribosome | | Descriptor: | (1S,2S,12bS,12cS)-2,4,5,7,12b,12c-hexahydro-1H-[1,3]dioxolo[4,5-j]pyrrolo[3,2,1-de]phenanthridine-1,2-diol, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4UEK
 
 | | Galactitol-1-phosphate 5-dehydrogenase from E. coli with Tris within the active site. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE, ZINC ION | | Authors: | Benavente, R, Esteban-Torres, M, Kohring, G.W, Cortes-Cabrera, A, Gago, F, Acebron, I, de las Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enantioselective Oxidation of Galactitol 1-Phosphate by Galactitol-1-Phosphate 5-Dehydrogenase from Escherichia Coli
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6TJF
 
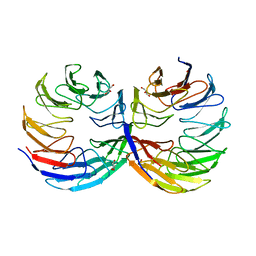 | | Crystal structure of the computationally designed Cake6 protein | | Descriptor: | Cake6, GLYCEROL | | Authors: | Mylemans, B, Laier, I, Voet, A.R.D, Noguchi, H. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
4UHV
 
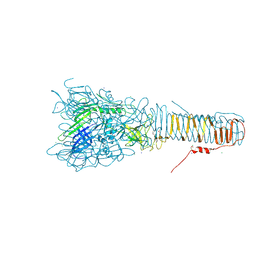 | | The structure of VgrG1, the needle tip of the bacterial Type VI Secretion System | | Descriptor: | CHLORIDE ION, SODIUM ION, VGRG1, ... | | Authors: | Spinola-Amilibia, M, Davo-Siguero, I, Ruiz, F.M, Santillana, E, Medrano, F.J, Romero, A. | | Deposit date: | 2015-03-25 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of Vgrg1 from Pseudomonas Aeruginosa, the Needle Tip of the Bacterial Type Vi Secretion System
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5XD9
 
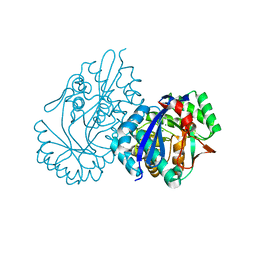 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily
Biochem. Biophys. Res. Commun., 491, 2017
|
|
6TJH
 
 | | Crystal structure of the computationally designed Cake9 protein | | Descriptor: | Cake9, GLYCEROL, SULFATE ION | | Authors: | Mylemans, B, Laier, I, Noguchi, H, Voet, A.R.D. | | Deposit date: | 2019-11-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural plasticity of a designer protein sheds light on beta-propeller protein evolution.
Febs J., 288, 2021
|
|
