6PNQ
 
 | | Crystal structure of the SS2A splice insert-containing neurexin-1 LNS2 domain in complex with neurexophilin-1 | | Descriptor: | Neurexin-1, Neurexophilin-1 | | Authors: | Wilson, S.C, White, K.I, Zhou, Q, Brunger, A.T. | | Deposit date: | 2019-07-02 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Structures of neurexophilin-neurexin complexes reveal a regulatory mechanism of alternative splicing.
Embo J., 38, 2019
|
|
2QCB
 
 | | T7-tagged full-length streptavidin complexed with ruthenium ligand | | Descriptor: | N-(4-{[(2-AMINOETHYL)AMINO]SULFONYL}PHENYL)-5-[(3AS,4S,6AR)-2-OXOHEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL]PENTANAMIDE-(1,2,3,4,5,6-ETA)-BENZENE-CHLORO-RUTHENIUM(III), Streptavidin | | Authors: | Le Trong, I, Creus, M, Pordea, A, Ward, T.R, Stenkamp, R.E. | | Deposit date: | 2007-06-19 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and designed evolution of an artificial transfer hydrogenase
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3L4D
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Leishmania infantum in complex with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Wawrzak, Z, Waterman, M.R. | | Deposit date: | 2009-12-19 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Substrate Preferences and Catalytic Parameters Determined by Structural Characteristics of Sterol 14{alpha}-Demethylase (CYP51) from Leishmania infantum.
J.Biol.Chem., 286, 2011
|
|
6YM0
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with CR3022 Fab (crystal form 1) | | Descriptor: | Spike glycoprotein, heavy chain, light chain | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Ginn, H.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.36 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
7Q1K
 
 | | Crystal structure of the native AA9A LPMO from Thermoascus aurantiacus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Yu, W, Mohsin, I, Li, D.C, Papageorgiou, A.C. | | Deposit date: | 2021-10-20 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Purification and Structural Characterization of the Auxiliary Activity 9 Native Lytic Polysaccharide Monooxygenase from Thermoascus aurantiacus and Identification of Its C1- and C4-Oxidized Reaction Products
Catalysts, 12, 2022
|
|
3KB8
 
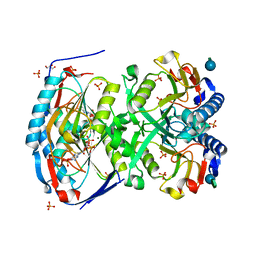 | | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP
To be Published
|
|
6YQG
 
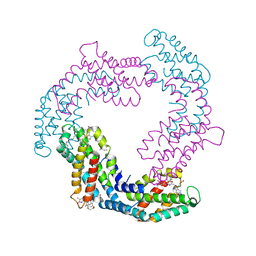 | | Crystal structure of native Phycocyanin in spacegroup P63 at 1.45 Angstroms. | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-17 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1D77
 
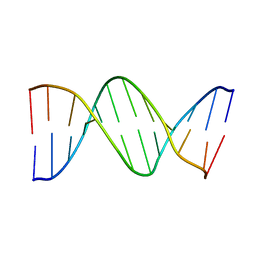 | |
4XCF
 
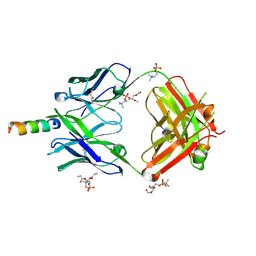 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 gp41; crystals cryoprotected with phosphatidylcholine (03:0 PC) | | Descriptor: | 4E10 Fab heavy chain, 4E10 Fab light chain, MODIFIED FRAGMENT OF HIV-1 GLYCOPROTEIN (GP41) INCLUDING THE MPER REGION 671-683, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
3ZUE
 
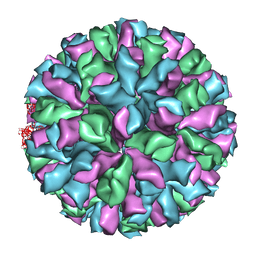 | | Rabbit Hemorrhagic Disease Virus (RHDV)capsid protein | | Descriptor: | CAPSID STRUCTURAL PROTEIN VP60 | | Authors: | Luque, D, Gonzalez, J.M, Gomez-Blanco, J, Marabini, R, Chichon, J, Mena, I, Angulo, I, Carrascosa, J.L, Verdaguer, N, Trus, B.L, Barcena, J, Caston, J.R. | | Deposit date: | 2011-07-18 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | Epitope Insertion at the N-Terminal Molecular Switch of the Rabbit Hemorrhagic Disease Virus T=3 Capsid Protein Leads to Larger T=4 Capsids.
J.Virol., 86, 2012
|
|
3KG2
 
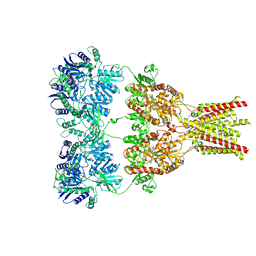 | | AMPA subtype ionotropic glutamate receptor in complex with competitive antagonist ZK 200775 | | Descriptor: | Glutamate receptor 2, beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sobolevsky, A.I, Rosconi, M.P, Gouaux, E. | | Deposit date: | 2009-10-28 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor.
Nature, 462, 2009
|
|
5CEZ
 
 | |
4Y1M
 
 | |
2QHF
 
 | | Mycobacterium tuberculosis Chorismate synthase in complex with NCA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Bruning, M, Bourenkov, G.P, Strizhov, N.I, Bartunik, H.D. | | Deposit date: | 2007-07-02 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mycobacterium tuberculosis Chorismate synthase in complex with NCA
To be Published
|
|
7VG5
 
 | | 10,5-methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1 with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Methenyltetrahydrofolate cyclohydrolase | | Authors: | Kim, S, Lee, S, Kim, I.-K, Seo, H, Kim, K.-J. | | Deposit date: | 2021-09-14 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into a molecular mechanism of methenyltetrahydrofolate cyclohydrolase from Methylobacterium extorquens AM1.
Int.J.Biol.Macromol., 202, 2022
|
|
4XIH
 
 | |
6Y58
 
 | | Binary complex of 14-3-3 sigma (C38N) with the Estrogen Related Receptor gamma (LBD) phosphopeptide | | Descriptor: | 14-3-3 protein sigma, Estrogen Related Receptor gamma phosphopeptide, MAGNESIUM ION | | Authors: | Somsen, B.A, Sijbesma, E, Leijten-van de Gevel, I.A, Ottmann, C. | | Deposit date: | 2020-02-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Fluorescence Anisotropy-Based Tethering for Discovery of Protein-Protein Interaction Stabilizers.
Acs Chem.Biol., 15, 2020
|
|
1D9I
 
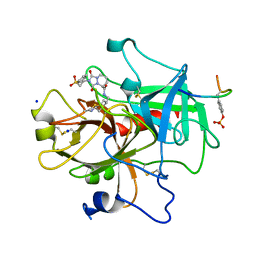 | | STRUCTURE OF THROMBIN COMPLEXED WITH SELECTIVE NON-ELECTOPHILIC INHIBITORS HAVING CYCLOHEXYL MOIETIES AT P1 | | Descriptor: | (5S)-N-[(trans-4-aminocyclohexyl)methyl]-1,3-dioxo-2-[2-(phenylsulfonyl)ethyl]-2,3,5,8-tetrahydro-1H-[1,2,4]triazolo[1,2-a]pyridazine-5-carboxamide, HIRUGEN, SODIUM ION, ... | | Authors: | Krishnan, R, Mochalkin, I, Arni, R, Tulinsky, A. | | Deposit date: | 1999-10-27 | | Release date: | 2000-10-30 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of thrombin complexed with selective non-electrophilic inhibitors having cyclohexyl moieties at P1.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3KGC
 
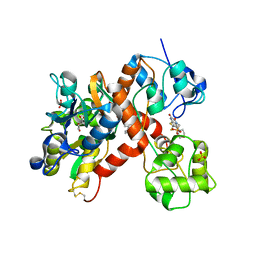 | | Isolated ligand binding domain dimer of GluA2 ionotropic glutamate receptor in complex with glutamate, LY 404187 and ZK 200775 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, N-[(2S)-2-(4'-cyanobiphenyl-4-yl)propyl]propane-2-sulfonamide, ... | | Authors: | Sobolevsky, A.I, Rosconi, M.P, Gouaux, E. | | Deposit date: | 2009-10-28 | | Release date: | 2009-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor
Nature, 462, 2009
|
|
3KHJ
 
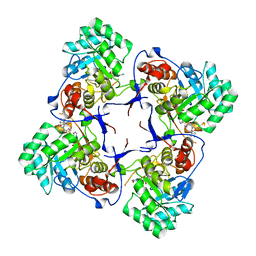 | |
3KIH
 
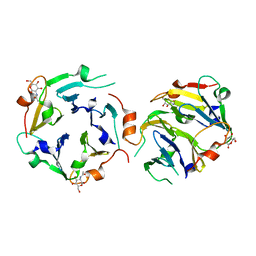 | | The crystal structures of two fragments truncated from 5-bladed beta-propeller lectin, tachylectin-2 (Lib2-D2-15) | | Descriptor: | 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 5-bladed beta-propeller lectin | | Authors: | Dym, O, Tawfik, D.S, Yadid, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-11-02 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Metamorphic proteins mediate evolutionary transitions of structure
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4XKD
 
 | |
7BJP
 
 | | The cryo-EM structure of vesivirus 2117, an adventitious agent and possible cause of haemorrhagic gastroenteritis in dogs. | | Descriptor: | Capsid protein | | Authors: | Sutherland, H, Conley, M.J, Emmott, E, Streetley, J, Goodfellow, I.G, Bhella, D. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | The Cryo-EM Structure of Vesivirus 2117 Highlights Functional Variations in Entry Pathways for Viruses in Different Clades of the Vesivirus Genus.
J.Virol., 95, 2021
|
|
3KEG
 
 | | X-ray Crystallographic Structure of a Y131F mutant of Pseudomonas Aeruginosa Azoreductase in complex with Methyl RED | | Descriptor: | 2-(4-DIMETHYLAMINOPHENYL)DIAZENYLBENZOIC ACID, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Wang, C.-J, Laurieri, N, Abuhammad, A, Lowe, E, Westwood, I, Ryan, A, Sim, E. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of tyrosine 131 in the active site of paAzoR1, an azoreductase with specificity for the inflammatory bowel disease prodrug balsalazide
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6P9D
 
 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with FAD - Yellow fraction | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent catabolic D-arginine dehydrogenase DauA, GLYCEROL | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
