1TPG
 
 | | F1-G MODULE PAIR RESIDUES 1-91 (C83S) OF TISSUE-TYPE PLASMINOGEN ACTIVATOR (T-PA) (NMR, 298K, PH2.95, REPRESENTATIVE STRUCTURE) | | Descriptor: | T-PLASMINOGEN ACTIVATOR F1-G | | Authors: | Smith, B.O, Downing, A.K, Driscoll, P.C, Dudgeon, T.J, Campbell, I.D. | | Deposit date: | 1995-06-14 | | Release date: | 1995-09-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure and backbone dynamics of the fibronectin type I and epidermal growth factor-like pair of modules of tissue-type plasminogen activator.
Structure, 3, 1995
|
|
8CIN
 
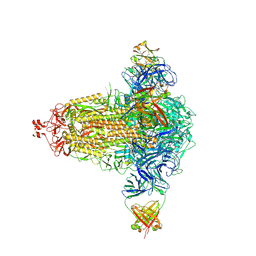 | | BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-5 fab HEAVY CHAIN, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2023-02-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8BZJ
 
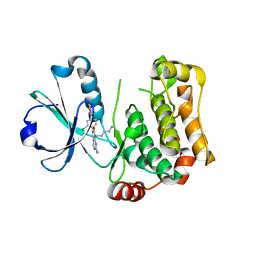 | | Human MST3 (STK24) kinase in complex with inhibitor MRLW5 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
5OJW
 
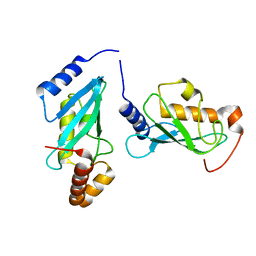 | | S. cerevisiae UBC13 - MMs2 complex | | Descriptor: | Ubiquitin-conjugating enzyme E2 13, Ubiquitin-conjugating enzyme variant MMS2 | | Authors: | Sharon, I, Rathi, R, Levin-Kravets, O, Attali, I, Prag, G. | | Deposit date: | 2017-07-24 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | S. cerevisiae UBC13 - MMs2 complex
To Be Published
|
|
7O06
 
 | |
4UOI
 
 | | Unexpected structure for the N-terminal domain of Hepatitis C virus envelope glycoprotein E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GENOME POLYPROTEIN | | Authors: | El Omari, K, Iourin, O, Kadlec, J, Harlos, K, Grimes, J.M, Stuart, D.I. | | Deposit date: | 2014-06-04 | | Release date: | 2014-08-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Unexpected Structure for the N-Terminal Domain of Hepatitis C Virus Envelope Glycoprotein E1
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8COA
 
 | | in situ Subtomogram average of Immature Rotavirus TLP spike | | Descriptor: | Intermediate capsid protein VP6, Outer capsid glycoprotein VP7, Outer capsid protein VP4 | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2023-02-27 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
7KM3
 
 | | Dodecameric Structure of the Chlamydia trachomatis Flavin Prenyltransferase UbiX Ortholog CT220 with FMN and DMAP | | Descriptor: | Dimethylallyl monophosphate, FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX | | Authors: | Nguyen, T, Nicely, N.I, Belaia-Martiniouk, A, Dunne, A.P, McCafferty, D.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Functional and structural validation of CT220 as the UbiX-like flavin prenyltransferase from Chlamydial menaquinone biosynthesis
To be published
|
|
1MJG
 
 | | CRYSTAL STRUCTURE OF BIFUNCTIONAL CARBON MONOXIDE DEHYDROGENASE/ACETYL-COA SYNTHASE(CODH/ACS) FROM MOORELLA THERMOACETICA (F. CLOSTRIDIUM THERMOACETICUM) | | Descriptor: | ACETATE ION, CARBON MONOXIDE DEHYDROGENASE BETA SUBUNIT, COPPER (I) ION, ... | | Authors: | Doukov, T.I, Iverson, T.M, Seravalli, J, Ragsdale, S.W, Drennan, C.L. | | Deposit date: | 2002-08-27 | | Release date: | 2003-01-28 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Ni-Fe-Cu center in a bifunctional carbon monoxide dehydrogenase/acetyl-CoA
synthase
Science, 298, 2002
|
|
5L3Q
 
 | | Structure of the GTPase heterodimer of human SRP54 and SRalpha | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wild, K, Segnitz, B, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
3KPD
 
 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | Authors: | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Chantar, M.L, Martinez-Cruz, L.A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
4UHL
 
 | | HUMAN STEROL 14-ALPHA DEMETHYLASE (CYP51) IN COMPLEX WITH VFV IN P1 SPACE GROUP | | Descriptor: | N-[(1R)-1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, I Lepesheva, G. | | Deposit date: | 2015-03-24 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human Sterol 14Alpha-Demethylase (Cyp51) as a Target for Anticancer Chemotherapy: Towards Structure-Aided Drug Design.
J.Lipid Res., 57, 2016
|
|
5J5P
 
 | | AMP-PNP-stabilized ATPase domain of topoisomerase IV from Streptococcus pneumoniae, complex type I | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*GP*C)-3'), DNA topoisomerase 4 subunit B, MAGNESIUM ION, ... | | Authors: | Laponogov, I, Pan, X.-S, Skamrova, G, Umrekar, T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2016-04-03 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Trapping of the transport-segment DNA by the ATPase domains of a type II topoisomerase.
Nat Commun, 9, 2018
|
|
6N5A
 
 | |
5J77
 
 | |
8XI6
 
 | | SARS-CoV-2 Omicron BQ.1.1 Variant Spike Protein Complexed with MO11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ishimaru, H, Nishimura, M, Shigematsu, H, Marini, M.I, Hasegawa, N, Takamiya, R, Iwata, S, Mori, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Epitopes of an antibody that neutralizes a wide range of SARS-CoV-2 variants in a conserved subdomain 1 of the spike protein.
J.Virol., 98, 2024
|
|
2QP2
 
 | | Structure of a MACPF/perforin-like protein | | Descriptor: | CALCIUM ION, Unknown protein | | Authors: | Rosado, C.J, Buckle, A.M, Law, R.H.P, Butcher, R.E, Kan, W.T, Bird, C.H, Ung, K, Browne, K.A, Baran, K, Bashtannyk-Puhalovich, T.A, Faux, N.G, Wong, W, Porter, C.J, Pike, R.N, Ellisdon, A.M, Pearce, M.C, Bottomley, S.P, Emsley, J, Smith, A.I, Rossjohn, J, Hartland, E.L, Voskoboinik, I, Trapani, J.A, Bird, P.I, Dunstone, M.A, Whisstock, J.C. | | Deposit date: | 2007-07-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A common fold mediates vertebrate defense and bacterial attack
Science, 317, 2007
|
|
5AIY
 
 | | R6 HUMAN INSULIN HEXAMER (SYMMETRIC), NMR, 'RED' SUBSTATE, AVERAGE STRUCTURE | | Descriptor: | PHENOL, PROTEIN (INSULIN) | | Authors: | O'Donoghue, S.I, Chang, X, Abseher, R, Nilges, M, Led, J.J. | | Deposit date: | 1998-12-29 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Unraveling the symmetry ambiguity in a hexamer: calculation of the R6 human insulin structure.
J.Biomol.NMR, 16, 2000
|
|
7KN8
 
 | | Crystal structure of the GH74 xyloglucanase from Xanthomonas campestris (Xcc1752) | | Descriptor: | 1,2-ETHANEDIOL, Cellulase, IODIDE ION, ... | | Authors: | Araujo, E.A, Vieira, P.S, Murakami, M.T, Polikarpov, I. | | Deposit date: | 2020-11-04 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Xyloglucan processing machinery in Xanthomonas pathogens and its role in the transcriptional activation of virulence factors
Nature Communications, 12, 2021
|
|
4UHI
 
 | | HUMAN STEROL 14-ALPHA DEMETHYLASE (CYP51) IN COMPLEX WITH VFV IN C121 SPACE GROUP | | Descriptor: | N-[(1R)-1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE | | Authors: | Hargrove, T.Y, Wawrzak, Z, I Lepesheva, G. | | Deposit date: | 2015-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Human Sterol 14Alpha-Demethylase (Cyp51) as a Target for Anticancer Chemotherapy: Towards Structure-Aided Drug Design.
J.Lipid Res., 57, 2016
|
|
6E7J
 
 | | HIV-1 wild type protease with GRL-042-17A, 3-phenylhexahydro-2h-cyclopenta[d]oxazol-2-one with a bicyclic oxazolidinone scaffold as the P2 ligand | | Descriptor: | (3aS,5R,6aR)-2-oxo-3-phenylhexahydro-2H-cyclopenta[d][1,3]oxazol-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2018-07-26 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design and Synthesis of Potent HIV-1 Protease Inhibitors Containing Bicyclic Oxazolidinone Scaffold as the P2 Ligands: Structure-Activity Studies and Biological and X-ray Structural Studies.
J. Med. Chem., 61, 2018
|
|
2Y8P
 
 | | Crystal Structure of an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase (MltE) from Escherichia coli | | Descriptor: | ENDO-TYPE MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE A | | Authors: | Artola-Recolons, C, Carrasco-Lopez, C, Llarrull, L.I, Kumarasiri, M, Lastochkin, E, Martinez-Ilarduya, I, Meindl, K, Uson, I, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | High-Resolution Crystal Structure of Mlte, an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase from Escherichia Coli.
Biochemistry, 50, 2011
|
|
7KQW
 
 | | Crystal structure of SARS-CoV-2 NSP3 macrodomain (C2 crystal form, methylated) | | Descriptor: | Non-structural protein 3 | | Authors: | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-11-17 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
1BX4
 
 | | STRUCTURE OF HUMAN ADENOSINE KINASE AT 1.50 ANGSTROMS | | Descriptor: | ADENOSINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mathews, I.I, Erion, M.D, Ealick, S.E. | | Deposit date: | 1998-10-13 | | Release date: | 1999-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of human adenosine kinase at 1.5 A resolution.
Biochemistry, 37, 1998
|
|
4X6G
 
 | |
