3OB6
 
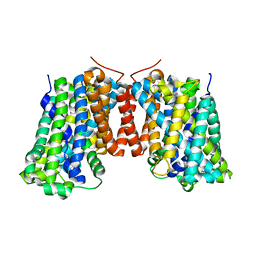 | | Structure of AdiC(N101A) in the open-to-out Arg+ bound conformation | | Descriptor: | ARGININE, AdiC arginine:agmatine antiporter | | Authors: | Carpena, X, Kowalczyk, L, Ratera, M, Valencia, E, Vazquez-lbar, J.L, Fita, I, Palacin, M. | | Deposit date: | 2010-08-06 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of substrate-induced permeation by an amino acid antiporter.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NOW
 
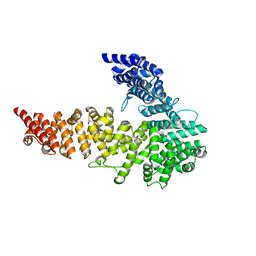 | | UNC-45 from Drosophila melanogaster | | Descriptor: | UNC-45 protein, SD10334p | | Authors: | Lee, C.F, Hauenstein, A.V, Fleming, J.K, Gasper, W.C, Engelke, V, Banumathi, S, Bernstein, S.I, Huxford, T. | | Deposit date: | 2010-06-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | X-ray Crystal Structure of the UCS Domain-Containing UNC-45 Myosin Chaperone from Drosophila melanogaster.
Structure, 19, 2011
|
|
3OC2
 
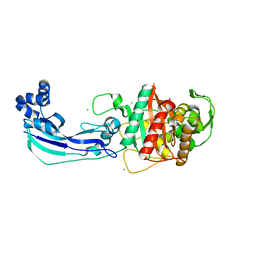 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Penicillin-binding protein 3 | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
3NVF
 
 | |
3NVG
 
 | |
3NR8
 
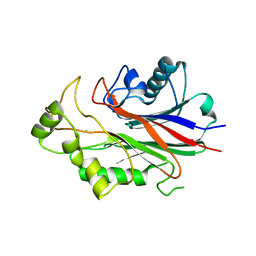 | | Crystal structure of human SHIP2 | | Descriptor: | CHLORIDE ION, Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases
Structure, 22, 2014
|
|
3R6K
 
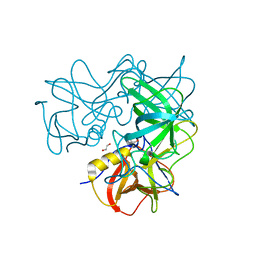 | | Crystal Structure of the Capsid P Domain from Norwalk Virus Strain Hiroshima/1999 in complex with HBGA type B (triglycan) | | Descriptor: | 1,2-ETHANEDIOL, VP1 protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Hansman, G.S, Biertumpfel, C, McLellan, J.S, Georgiev, I, Chen, L, Zhou, T, Katayama, K, Kwong, P.D. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
3R2T
 
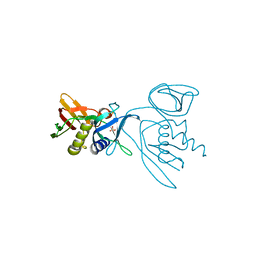 | | 2.2 Angstrom Resolution Crystal Structure of Superantigen-like Protein from Staphylococcus aureus subsp. aureus NCTC 8325. | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Kiryukhina, O, Falugi, F, Bottomley, M, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-14 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure of Superantigen-like Protein from Staphylococcus aureus subsp. aureus NCTC 8325.
TO BE PUBLISHED
|
|
3R72
 
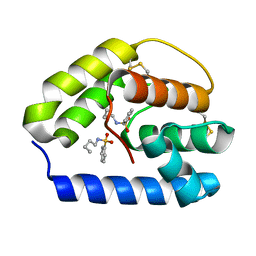 | | Apis mellifera odorant binding protein 5 | | Descriptor: | N-BUTYL-BENZENESULFONAMIDE, Odorant binding protein ASP5 | | Authors: | Spinelli, S, Iovinella, I, Lagarde, A, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-03-22 | | Release date: | 2012-04-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of Apis mellifera odorant binding protein 5
To be Published
|
|
3RK4
 
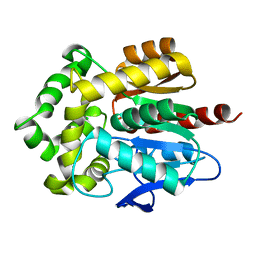 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2011-04-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3R0Y
 
 | | Crystal Structures of Multidrug-resistant HIV-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors | | Descriptor: | Multidrug-resistant clinical isolate 769 HIV-1 Protease, N-[(2S)-1-{[(2S,3S)-3-hydroxy-5-oxo-5-{[(2R)-1-oxo-3-phenyl-1-(prop-2-yn-1-ylamino)propan-2-yl]amino}-1-phenylpentan-2-yl]amino}-3-methyl-1-oxobutan-2-yl]pyridine-2-carboxamide | | Authors: | Yedidi, R.S, Gupta, D, Liu, Z, Brunzelle, J, Kovari, I.A, Woster, P.M, Kovari, L.C. | | Deposit date: | 2011-03-09 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of multidrug-resistant HIV-1 protease in complex with two potent anti-malarial compounds.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
3RJZ
 
 | | X-ray crystal structure of the putative n-type atp pyrophosphatase from pyrococcus furiosus, the northeast structural genomics target pfr23 | | Descriptor: | N-type ATP pyrophosphatase superfamily | | Authors: | Forouhar, F, Lee, I, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-16 | | Release date: | 2011-05-11 | | Last modified: | 2012-01-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A large conformational change in the putative ATP pyrophosphatase PF0828 induced by ATP binding.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3RE4
 
 | |
5UAB
 
 | |
5UAD
 
 | |
3INW
 
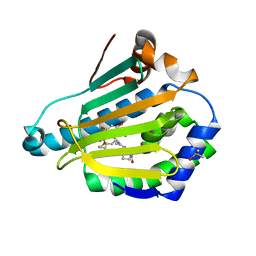 | | HSP90 N-TERMINAL DOMAIN with pochoxime A | | Descriptor: | (5E,9E,11E)-13-chloro-14,16-dihydroxy-3,4,7,8-tetrahydro-1H-2-benzoxacyclotetradecine-1,11(12H)-dione 11-[O-(2-oxo-2-piperidin-1-ylethyl)oxime], DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha | | Authors: | Korndoerfer, I.P. | | Deposit date: | 2009-08-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibition of HSP90 with pochoximes: SAR and structure-based insights.
Chembiochem, 10, 2009
|
|
3INX
 
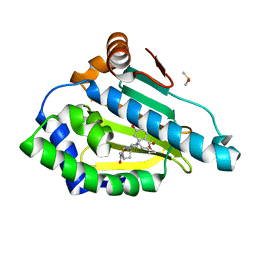 | | HSP90 N-TERMINAL DOMAIN with pochoxime B | | Descriptor: | (5E,9E,11E)-14,16-dihydroxy-3,4,7,8-tetrahydro-1H-2-benzoxacyclotetradecine-1,11(12H)-dione 11-[O-(2-oxo-2-piperidin-1-ylethyl)oxime], DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha | | Authors: | Korndoerfer, I.P. | | Deposit date: | 2009-08-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inhibition of HSP90 with pochoximes: SAR and structure-based insights.
Chembiochem, 10, 2009
|
|
3HNX
 
 | |
3I1G
 
 | |
2J8H
 
 | | Structure of the immunoglobulin tandem repeat A168-A169 of titin | | Descriptor: | GLYCEROL, TITIN | | Authors: | Mueller, S, Lange, S, Kursula, I, Gautel, M, Wilmanns, M. | | Deposit date: | 2006-10-25 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rigid Conformation of an Immunoglobulin Domain Tandem Repeat in the A-Band of the Elastic Muscle Protein Titin
J.Mol.Biol., 371, 2007
|
|
2HVX
 
 | | Discovery of Potent, Orally Active, Nonpeptide Inhibitors of Human Mast Cell Chymase by Using Structure-Based Drug Design | | Descriptor: | Chymase, [(1S)-1-(5-CHLORO-1-BENZOTHIEN-3-YL)-2-(2-NAPHTHYLAMINO)-2-OXOETHYL]PHOSPHONIC ACID | | Authors: | Greco, M.N, Hawkins, M.J, Powell, E.T, Almond, H.R, de Garavilla, L, Wang, Y, Minor, L.A, Wells, G.I, Di Cera, E, Cantwell, A.M, Savvides, S.N, Damiano, B.P, Maryanoff, B.E. | | Deposit date: | 2006-07-31 | | Release date: | 2007-06-12 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of potent, selective, orally active, nonpeptide inhibitors of human mast cell chymase.
J.Med.Chem., 50, 2007
|
|
5COL
 
 | |
2K8V
 
 | | Solution structure of Oxidised ERp18 | | Descriptor: | Thioredoxin domain-containing protein 12 | | Authors: | Rowe, M.L, Alanen, H.I, Ruddock, L.W, Kelly, G, Schmidt, J.M, Williamson, R.A, Howard, M.J. | | Deposit date: | 2008-09-25 | | Release date: | 2009-06-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ERp18, a small endoplasmic reticulum resident oxidoreductase .
Biochemistry, 48, 2009
|
|
8CLN
 
 | | Zearalenone lactonase from Streptomyces coelicoflavus, SeMet derivative for SAD phasing | | Descriptor: | Hydrolase | | Authors: | Puehringer, D, Grishkovskaya, I, Mlynek, G, Kostan, J. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
3DA0
 
 | | Crystal structure of a cleaved form of a chimeric receptor binding protein from Lactococcal phages subspecies TP901-1 and p2 | | Descriptor: | Cleaved chimeric receptor binding protein from bacteriophages TP901-1 and p2 | | Authors: | Siponen, M.I, Blangy, S, Spinelli, S, Vera, L, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a chimeric receptor binding protein constructed from two lactococcal phages.
J.Bacteriol., 191, 2009
|
|
