1GXO
 
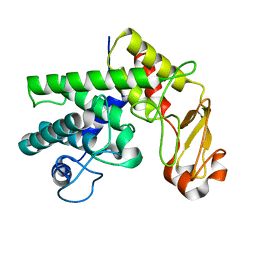 | | Mutant D189A of Family 10 polysaccharide lyase from Cellvibrio cellulosa in complex with trigalaturonic acid | | Descriptor: | CALCIUM ION, PECTATE LYASE, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Charnock, S.J, Brown, I.E, Turkenburg, J.P, Black, G.W, Davies, G.J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Convergent Evolution Sheds Light on the Anti-Beta-Elimination Mechanism Common to Family 1 and 10 Polysaccharide Lyases
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7MLF
 
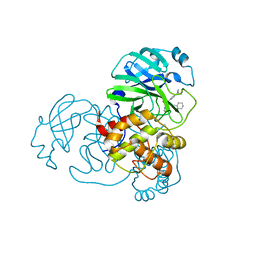 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C7 | | Descriptor: | 3C-like proteinase, N-(4-tert-butylphenyl)-2-chloro-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]acetamide | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
3OU1
 
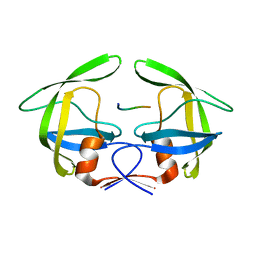 | | MDR769 HIV-1 protease complexed with RH/IN hepta-peptide | | Descriptor: | MDR HIV-1 protease, RH/IN substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
7MLG
 
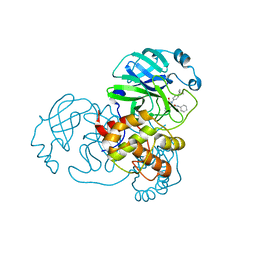 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C63 | | Descriptor: | (2R)-2-[(4-tert-butylphenyl)(ethanesulfonyl)amino]-N-cyclohexyl-2-(pyridin-3-yl)acetamide, 3C-like proteinase | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
2WTD
 
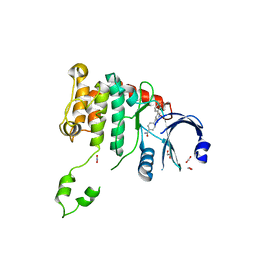 | | Crystal structure of Chk2 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-AMINO-5-(1,3-BENZODIOXOL-4-YL)PYRIDIN-3-YL]BENZAMIDE, NITRATE ION, ... | | Authors: | Hilton, S, Naud, S, Caldwell, J.J, Boxall, K, Burns, S, Anderson, V.E, Antoni, L, Allen, C.E, Pearl, L.H, Oliver, A.W, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2009-09-15 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification and Characterisation of 2-Aminopyridine Inhibitors of Checkpoint Kinase 2
Bioorg.Med.Chem., 18, 2010
|
|
4LDO
 
 | | Structure of beta2 adrenoceptor bound to adrenaline and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Camelid Antibody Fragment, L-EPINEPHRINE, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
1OLZ
 
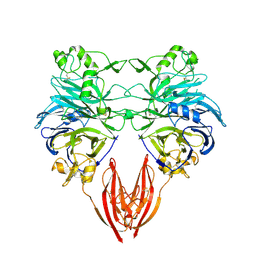 | | The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D | | Descriptor: | SEMAPHORIN 4D | | Authors: | Love, C.A, Harlos, K, Mavaddat, N, Davis, S.J, Stuart, D.I, Jones, E.Y, Esnouf, R.M. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-11 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Nat.Struct.Biol., 10, 2003
|
|
1Z1D
 
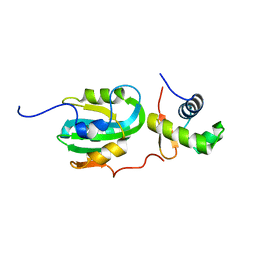 | | Structural Model for the interaction between RPA32 C-terminal domain and SV40 T antigen origin binding domain. | | Descriptor: | Large T antigen, Replication protein A 32 kDa subunit | | Authors: | Arunkumar, A.I, Klimovich, V, Jiang, X, Ott, R.D, Mizoue, L, Fanning, E, Chazin, W.J. | | Deposit date: | 2005-03-03 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Insights into hRPA32 C-terminal domain--mediated assembly of the simian virus 40 replisome.
Nat.Struct.Mol.Biol., 12, 2005
|
|
4LJ8
 
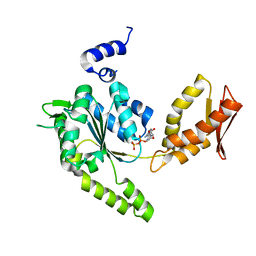 | | ClpB NBD2 R621Q from T. thermophilus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein ClpB | | Authors: | Zeymer, C, Barends, T.R.M, Werbeck, N.D, Schlichting, I, Reinstein, J. | | Deposit date: | 2013-07-04 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: a structure-based mechanistic dissection of a molecular motor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QX3
 
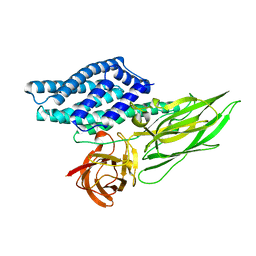 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2WIZ
 
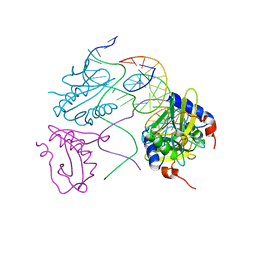 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | ARCHAEAL HJC, HALF-JUNCTION | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
1QPU
 
 | | SOLUTION STRUCTURE OF OXIDIZED ESCHERICHIA COLI CYTOCHROME B562 | | Descriptor: | CYTOCHROME B562, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnesano, F, Banci, L, Bertini, I, Faraone-Mennella, J, Rosato, A, Barker, P.D, Fersht, A.R. | | Deposit date: | 1999-05-30 | | Release date: | 1999-06-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of oxidized Escherichia coli cytochrome b562.
Biochemistry, 38, 1999
|
|
4H01
 
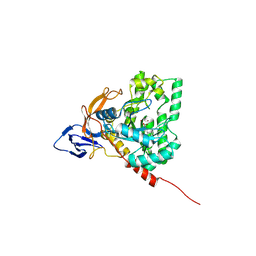 | | The crystal structure of di-Zn dihydropyrimidinase from Tetraodon nigroviridis | | Descriptor: | ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2012-09-06 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lysine Carboxylation: Metal and Structural Requirements for Post-translational Modification
To be Published
|
|
4QSU
 
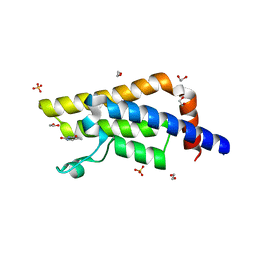 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) in complex with thymine | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-06 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based approaches towards identification of fragments for the low-druggability ATAD2 bromodomain
MedChemComm, 5, 2014
|
|
4R0R
 
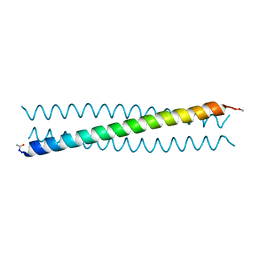 | | Ebolavirus GP Prehairpin Intermediate Mimic | | Descriptor: | eboIZN21 | | Authors: | Clinton, T.R, Weinstock, M.T, Jacobsen, M.T, Szabo-Fresnais, N, Pandya, M.J, Whitby, F.G, Herbert, A.S, Prugar, L.I, McKinnon, R, Hill, C.P, Welch, B.D, Dye, J.M, Eckert, D.M, Kay, M.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and characterization of ebolavirus GP prehairpin intermediate mimics as drug targets.
Protein Sci., 24, 2015
|
|
1ZL9
 
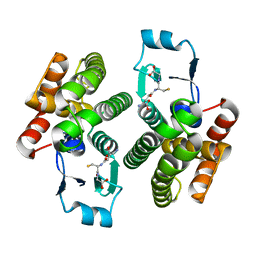 | | Crystal Structure of a major nematode C.elegans specific GST (CE01613) | | Descriptor: | GLUTATHIONE, glutathione S-transferase 5 | | Authors: | Kriksunov, I.A, Liu, Q, Schuller, D.J, Campbell, A.M, Barrett, J, Brophy, P.M, Hao, Q. | | Deposit date: | 2005-05-05 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of a major nematode C.elegans specific GST (CE01613)
To be Published
|
|
4GZW
 
 | | N2 neuraminidase D151G mutant of A/Tanzania/205/2010 H3N2 in complex with avian sialic acid receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic Acid receptors.
J.Virol., 86, 2012
|
|
2WI2
 
 | | Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone | | Descriptor: | 4-METHYL-6-(METHYLSULFANYL)-1,3,5-TRIAZIN-2-AMINE, DIMETHYL SULFOXIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Barril, X, Borgognoni, J, Chene, P, Davies, N.G.M, Davis, B, Drysdale, M.J, Dymock, B, Eccles, S.A, Garcia-Echeverria, C, Fromont, C, Hayes, A, Hubbard, R.E, Jordan, A.M, Rugaard-Jensen, M, Massey, A, Merret, A, Padfield, A, Parsons, R, Radimerski, T, Raynaud, F.I, Robertson, A, Roughley, S.D, Schoepfer, J, Simmonite, H, Surgenor, A, Valenti, M, Walls, S, Webb, P, Wood, M, Workman, P, Wright, L.M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Combining Hit Identification Strategies: Fragment- Based and in Silico Approaches to Orally Active 2-Aminothieno[2,3-D]Pyrimidine Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 52, 2009
|
|
2WJ0
 
 | | Crystal structures of Holliday junction resolvases from Archaeoglobus fulgidus bound to DNA substrate | | Descriptor: | ARCHAEAL HJC, COBALT HEXAMMINE(III), HALF-JUNCTION | | Authors: | Carolis, C, Koehler, C, Sauter, C, Basquin, J, Suck, D, Toeroe, I. | | Deposit date: | 2009-05-18 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Holliday Junction Resolvases from Archaeoglobus Fulgidus Bound to DNA Substrate
To be Published
|
|
2WK4
 
 | | Dimeric structure of D347G D348G mutant of the sapporovirus RNA dependent RNA polymerase | | Descriptor: | GLYCEROL, PROTEASE-POLYMERASE P70 | | Authors: | Fullerton, S.W.B, Robel, I, Schuldt, L, Gebhardt, J, Tucker, P.A, Rohayem, J. | | Deposit date: | 2009-06-05 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Dimeric Structure of D347G D348G Mutant of the Sapporovirus Sapporovirus RNA Dependent RNA Polymerase
To be Published
|
|
2AF2
 
 | | Solution structure of disulfide reduced and copper depleted Human Superoxide Dismutase | | Descriptor: | Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Banci, L, Bertini, I, Cantini, F, D'Amelio, N, Gaggelli, E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-07-25 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Human SOD1 before harboring the catalytic metal: solution structure of copper-depleted, disulfide-reduced form
J.Biol.Chem., 281, 2006
|
|
1F94
 
 | | THE 0.97 RESOLUTION STRUCTURE OF BUCANDIN, A NOVEL TOXIN ISOLATED FROM THE MALAYAN KRAIT | | Descriptor: | BUCANDIN | | Authors: | Kuhn, P, Deacon, A.M, Comoso, S, Rajaseger, G, Kini, R.M, Uson, I, Kolatkar, P.R. | | Deposit date: | 2000-07-06 | | Release date: | 2000-07-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | The atomic resolution structure of bucandin, a novel toxin isolated from the Malayan krait, determined by direct methods.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1Q6L
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound L-threonohydroxamate 4-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-THREONOHYDROXAMATE 4-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
1Q6A
 
 | | Solution Structure of the C-terminal Domain of Thermosynechococcus elongatus KaiA (ThKaiA180C); Averaged Minimized Structure | | Descriptor: | Circadian clock protein KaiA homolog | | Authors: | Vakonakis, I, Sun, J, Holzenburg, A, Golden, S.S, LiWang, A.C. | | Deposit date: | 2003-08-13 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: Implications for KaiA-KaiC interaction
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4LNW
 
 | | Crystal structure of TR-alpha bound to T3 in a second site | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor alpha | | Authors: | Puhl, A.C, Aparicio, R, Polikarpov, I. | | Deposit date: | 2013-07-12 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a new hormone-binding site on the surface of thyroid hormone receptor.
Mol.Endocrinol., 28, 2014
|
|
