2Q0V
 
 | | Crystal structure of ubiquitin conjugating enzyme E2, putative, from Plasmodium falciparum | | Descriptor: | PHOSPHATE ION, Ubiquitin-conjugating enzyme E2, putative | | Authors: | Wernimont, A.K, Lew, J, Hassanali, A, Lin, L, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-22 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ubiquitin conjugating enzyme E2, putative, from Plasmodium falciparum.
To be Published
|
|
5ANL
 
 | | Crystal structure of VPS34 in complex with (2S)-8-((3R)-3- Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3, 4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one, processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | (8S)-2-(morpholin-4-yl)-9-[2-(propan-2-yloxy)ethyl]-8-(trifluoromethyl)-6,7,8,9-tetrahydro-4H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3 | | Authors: | Zander, U, Hoffmann, G, Mathieu, M, Marquette, J.-P, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
1MRL
 
 | | Crystal structure of streptogramin A acetyltransferase with dalfopristin | | Descriptor: | 5-(2-DIETHYLAMINO-ETHANESULFONYL)-21-HYDROXY-10-ISOPROPYL-11,19-DIMETHYL-9,26-DIOXA-3,15,28-TRIAZA-TRICYCLO[23.2.1.00,255]OCTACOSA-1(27),12,17,19,25(28)-PENTAENE-2,8,14,23-TETRAONE, Streptogramin A acetyltransferase | | Authors: | Kehoe, L.E, Snidwongse, J, Courvalin, P, Rafferty, J.B, Murray, I.A. | | Deposit date: | 2002-09-18 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Synercid (Quinupristin-Dalfopristin) Resistance in Gram-positive Bacterial Pathogens
J.Biol.Chem., 278, 2003
|
|
1TQG
 
 | | CheA phosphotransferase domain from Thermotoga maritima | | Descriptor: | Chemotaxis protein cheA | | Authors: | Quezada, C.M, Gradinaru, C, Simon, M.I, Bilwes, A.M, Crane, B.R. | | Deposit date: | 2004-06-17 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Helical Shifts Generate Two Distinct Conformers in the Atomic Resolution Structure of the CheA Phosphotransferase Domain from Thermotoga maritima.
J.Mol.Biol., 341, 2004
|
|
2FCA
 
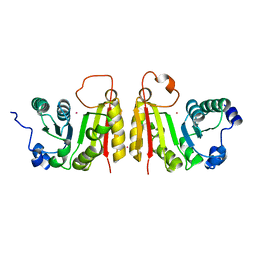 | | The structure of BsTrmB | | Descriptor: | POTASSIUM ION, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Zegers, I, Van Vliet, F, Bujnicki, J, Kosinski, J, Gigot, D, Droogmans, L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase.
Nucleic Acids Res., 34, 2006
|
|
6FX4
 
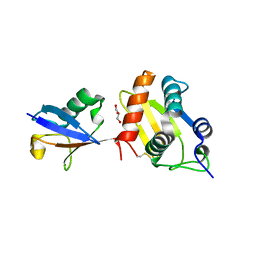 | |
5UNK
 
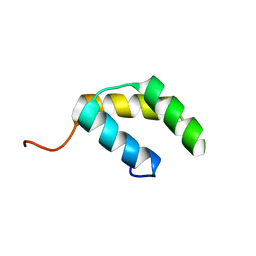 | |
2Q9P
 
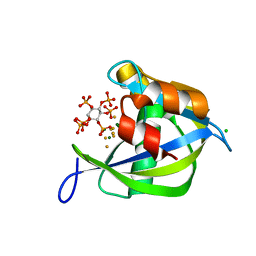 | | Human diphosphoinositol polyphosphate phosphohydrolase 1, Mg-F complex | | Descriptor: | CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, FLUORIDE ION, ... | | Authors: | Thorsell, A.G, Busam, R, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nordlund, P, Nyman, T, Ogg, D, Sagemark, J, Sundstrom, M, Van den Berg, S, Weigelt, J, Welin, M, Persson, C, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-13 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of human diphosphoinositol phosphatase 1.
Proteins, 77, 2009
|
|
3T6Y
 
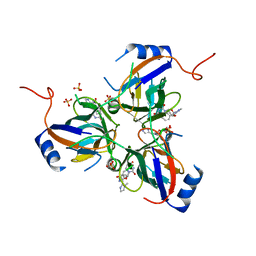 | | 5'-Diphenyl Nucleoside Inhibitors of Plasmodium falciparum dUTPase | | Descriptor: | 2',5'-dideoxy-5'-{[(R)-(1-methyl-1H-imidazol-2-yl)(phenyl)methyl]amino}uridine, Deoxyuridine 5'-triphosphate nucleotidohydrolase, putative, ... | | Authors: | Hampton, S.E, Baragana, B, Schipani, A, Bosch-Navarrete, C, Musso-Buendia, A, Recio, E, Kaiser, M, Whittingham, J.L, Roberts, S.M, Shevtsov, M, Brannigan, J.A, Kahnberg, P, Brun, R, Wilson, K.S, Gonzalez-Pacanowska, D, Johansson, N.G, Gilbert, I.H. | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis, and evaluation of 5'-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase.
Chemmedchem, 6, 2011
|
|
6GBR
 
 | | Crystal Structure of the oligomerization domain of VP35 from Reston virus, mercury derivative | | Descriptor: | MERCURIBENZOIC ACID, Polymerase cofactor VP35 | | Authors: | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
1N2S
 
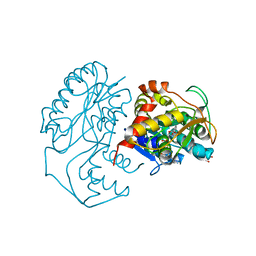 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-L-LYXO-4-HEXULOSE REDUCTASE (RMLD) IN COMPLEX WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Blankenfeldt, W, Kerr, I.D, Giraud, M.F, Mcmiken, H.J, Leonard, G.A, Whitfield, C, Messner, P, Graninger, M, Naismith, J.H. | | Deposit date: | 2002-10-24 | | Release date: | 2002-11-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variation on a Theme of SDR. dTDP-6-Deoxy-L- lyxo-4-Hexulose Reductase (RmlD) Shows a New Mg(2+)-Dependent Dimerization Mode
Structure, 10, 2002
|
|
6G0C
 
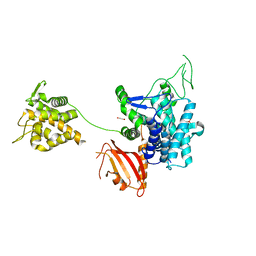 | | Crystal structure of SdeA catalytic core | | Descriptor: | 1,2-ETHANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Ubiquitinating/deubiquitinating enzyme SdeA | | Authors: | Kalayil, S, Bhogaraju, S, Basquin, J, Dikic, I. | | Deposit date: | 2018-03-17 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination.
Nature, 557, 2018
|
|
2PXA
 
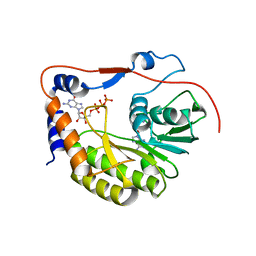 | | Crystal structure of the Murray Valley Encephalitis Virus NS5 2'-O Methyltransferase domain in complex with SAH and GTPG | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein [Contains: Capsid protein C (Core protein); Envelope protein M (Matrix protein); Major envelope protein E; Non-structural protein 1 (NS1); Non-structural protein 2A (NS2A); Flavivirin protease NS2B regulatory subunit; Flavivirin protease NS3 catalytic subunit; Non-structural protein 4A (NS4A); Non-structural protein 4B (NS4B); RNA-directed RNA polymerase (EC 2.7.7.48) (NS5)], S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Assenberg, R, Ren, J, Verma, A, Walter, T.S, Alderton, D, Hurrelbrink, R.J, Fuller, S.D, Owens, R.J, Stuart, D.I, Grimes, J.M, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2007-05-14 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Murray Valley encephalitis virus NS5 methyltransferase domain in complex with cap analogues.
J.Gen.Virol., 88, 2007
|
|
5B7D
 
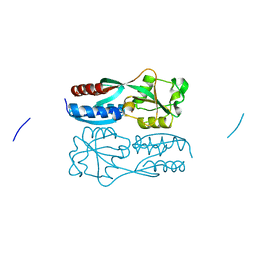 | |
1MT4
 
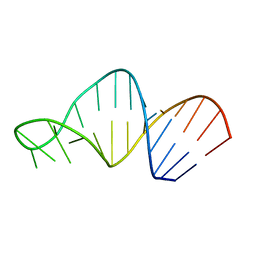 | | Structure of 23S ribosomal RNA hairpin 35 | | Descriptor: | 23S ribosomal Hairpin 35 | | Authors: | Lebars, I, Yoshizawa, S, Stenholm, A.R, Guittet, E, Douthwaite, S, Fourmy, D. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of 23S rRNA hairpin 35 and its interaction with the tylosin-resistance methyltransferase RlmAII
Embo J., 22, 2003
|
|
3SRH
 
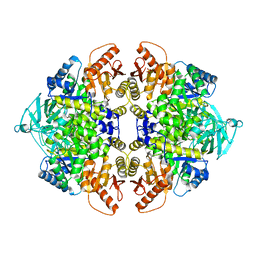 | | Human M2 pyruvate kinase | | Descriptor: | PHOSPHATE ION, Pyruvate kinase isozymes M1/M2 | | Authors: | Morgan, H.P, O'Reilly, F, Palmer, R, McNae, I.W, Nowicki, M.W, Wear, M.A, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosetric regulation of M2 pyruvate kinase.
To be Published
|
|
6FSF
 
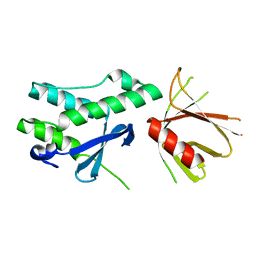 | | Crystal structure of the tandem PX-PH-domains of Bem3 from Saccharomyces cerevisiae | | Descriptor: | GTPase-activating protein BEM3 | | Authors: | Ali, I, Eu, S, Koch, D, Bleimling, N, Goody, R.S, Mueller, M.P. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the tandem PX-PH domains of Bem3 from Saccharomyces cerevisiae.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2Q9M
 
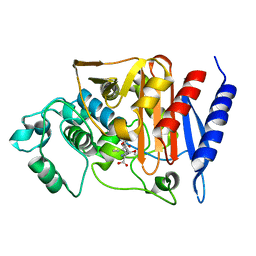 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | Descriptor: | (1R,4S,7AS)-1-(1-FORMYLPROP-1-EN-1-YL)-4-METHOXY-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
1MYT
 
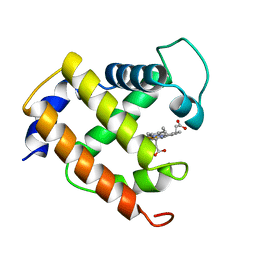 | | CRYSTAL STRUCTURE TO 1.74 ANGSTROMS RESOLUTION OF METMYOGLOBIN FROM YELLOWFIN TUNA (THUNNUS ALBACARES): AN EXAMPLE OF A MYOGLOBIN LACKING THE D HELIX | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Birnbaum, G.I, Evans, S.V, Przybylska, M, Rose, D.R. | | Deposit date: | 1991-05-06 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | 1.70 A resolution structure of myoglobin from yellowfin tuna. An example of a myoglobin lacking the D helix.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1MW2
 
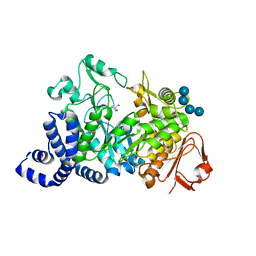 | | Amylosucrase soaked with 100mM sucrose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, amylosucrase, ... | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
6FSL
 
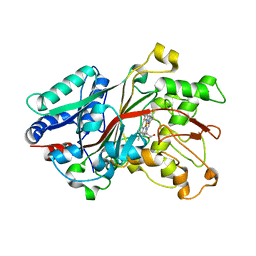 | |
1N1P
 
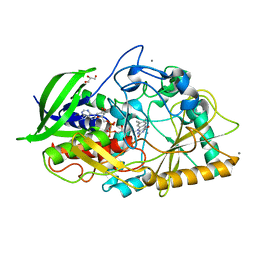 | |
5VRT
 
 | | Nonheme Iron Replacement in a Biosynthetic Nitric Oxide Reductase Model Performing O2 Reduction to Water: Co-bound FeBMb | | Descriptor: | COBALT (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Reed, J, Shi, Y, Zhu, Q, Chakraborty, S, Mirs, E.N, Petrik, I.D, Bhagi-Damodaran, A, Ross, M, Moenne-Loccoz, P, Zhang, Y, Lu, Y. | | Deposit date: | 2017-05-11 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Manganese and Cobalt in the Nonheme-Metal-Binding Site of a Biosynthetic Model of Heme-Copper Oxidase Superfamily Confer Oxidase Activity through Redox-Inactive Mechanism.
J. Am. Chem. Soc., 139, 2017
|
|
5VNU
 
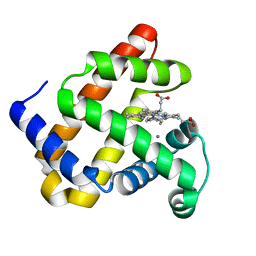 | | Nonheme Iron Replacement in a Biosynthetic Nitric Oxide Reductase Model Performing O2 Reduction to Water: Mn-bound FeBMb | | Descriptor: | MANGANESE (II) ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Reed, J, Shi, Y, Zhu, Q, Chakraborty, S, Mirs, E.N, Petrik, I.D, Bhagi-Damodaran, A, Ross, M, Moenne-Loccoz, P, Zhang, Y, Lu, Y. | | Deposit date: | 2017-05-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Manganese and Cobalt in the Nonheme-Metal-Binding Site of a Biosynthetic Model of Heme-Copper Oxidase Superfamily Confer Oxidase Activity through Redox-Inactive Mechanism.
J. Am. Chem. Soc., 139, 2017
|
|
1MW3
 
 | | Amylosucrase soaked with 1M sucrose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Skov, L.K, Mirza, O, Sprogoe, D, Dar, I, Remaud-Simeon, M, Albenne, C, Monsan, P, Gajhede, M. | | Deposit date: | 2002-09-27 | | Release date: | 2002-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide and Sucrose Complexes of Amylosucrase. STRUCTURAL IMPLICATIONS FOR THE POLYMERASE ACTIVITY
J.BIOL.CHEM., 277, 2002
|
|
