3V3X
 
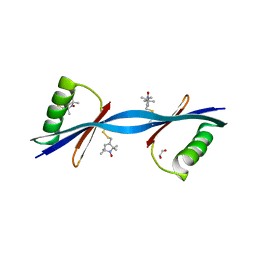 | | Nitroxide Spin Labels in Protein GB1: N8/K28 Double Mutant | | Descriptor: | ACETATE ION, GLYCEROL, Immunoglobulin G-binding protein G, ... | | Authors: | Cunningham, T.F, McGoff, M.S, Sengupta, I, Jaroniec, C.P, Horne, W.S, Saxena, S.K. | | Deposit date: | 2011-12-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a protein spin-label in a solvent-exposed beta-sheet and comparison with DEER spectroscopy.
Biochemistry, 51, 2012
|
|
5D6A
 
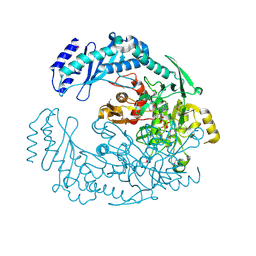 | | 2.7 Angstrom Crystal Structure of ABC transporter ATPase from Vibrio vulnificus in Complex with Adenylyl-imidodiphosphate (AMP-PNP) | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Predicted ATPase of the ABC class, SODIUM ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-11 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom Crystal Structure of ABC transporter ATPase from Vibrio vulnificus in Complex with Adenylyl-imidodiphosphate (AMP-PNP)
To Be Published
|
|
5W6L
 
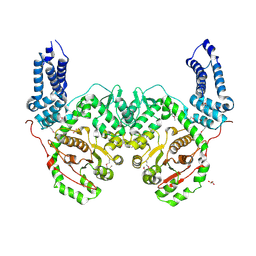 | | Crystal Structure of RRSP, a MARTX Toxin Effector Domain from Vibrio vulnificus CMCP6 | | Descriptor: | CHLORIDE ION, GLYCEROL, RTX repeat-containing cytotoxin, ... | | Authors: | Minasov, G, Wawrzak, Z, Biancucci, M, Shuvalova, L, Dubrovska, I, Satchell, K.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The bacterial Ras/Rap1 site-specific endopeptidase RRSP cleaves Ras through an atypical mechanism to disrupt Ras-ERK signaling.
Sci Signal, 11, 2018
|
|
1JHQ
 
 | | Three-dimensional Structure of CobT in Complex with Reaction Products of 5-methoxybenzimidazole and NaMN | | Descriptor: | N1-(5'-PHOSPHO-ALPHA-RIBOSYL)-5-METHOXYBENZIMIDAZOLE, NICOTINIC ACID, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
1JID
 
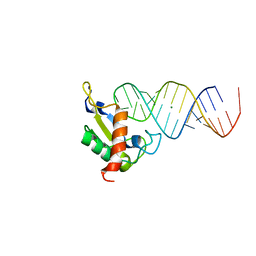 | | Human SRP19 in complex with helix 6 of Human SRP RNA | | Descriptor: | HELIX 6 OF HUMAN SRP RNA, MAGNESIUM ION, SIGNAL RECOGNITION PARTICLE 19 KDA PROTEIN | | Authors: | Wild, K, Sinning, I, Cusack, S. | | Deposit date: | 2001-07-02 | | Release date: | 2001-10-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an early protein-RNA assembly complex of the signal recognition particle.
Science, 294, 2001
|
|
3V6F
 
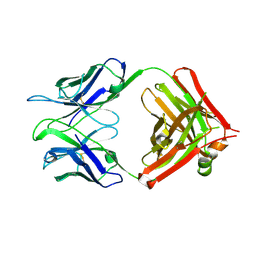 | | Crystal Structure of an anti-HBV e-antigen monoclonal Fab fragment (e6), unbound | | Descriptor: | Fab e6 Heavy Chain, Fab e6 Light Chain | | Authors: | Dimattia, M.A, Watts, N.R, Stahl, S.J, Grimes, J.M, Steven, A.C, Stuart, D.I, Wingfield, P.T. | | Deposit date: | 2011-12-19 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein.
Structure, 21, 2013
|
|
5DD8
 
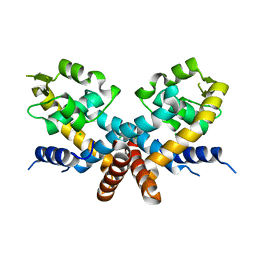 | | The Crystal structure of HucR mutant (HucR-E48Q) from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, Transcriptional regulator, MarR family | | Authors: | Deochand, D.K, Perera, I.C, Crochet, R.B, Gilbert, N.C, Newcomer, M.E, Grove, A. | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Histidine switch controlling pH-dependent protein folding and DNA binding in a transcription factor at the core of synthetic network devices.
Mol Biosyst, 12, 2016
|
|
3K1B
 
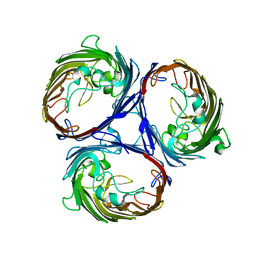 | | Structure of OmpF porin | | Descriptor: | Outer membrane protein F | | Authors: | Kefala, G, Ahn, C, Krupa, M, Maslennikov, I, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-09-26 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.39 Å) | | Cite: | Structures of the OmpF porin crystallized in the presence of foscholine-12.
Protein Sci., 19, 2010
|
|
1JCL
 
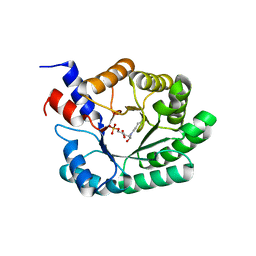 | | OBSERVATION OF COVALENT INTERMEDIATES IN AN ENZYME MECHANISM AT ATOMIC RESOLUTION | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, DEOXYRIBOSE-PHOSPHATE ALDOLASE | | Authors: | Heine, A, DeSantis, G, Luz, J.G, Mitchell, M, Wong, C.-H, Wilson, I.A. | | Deposit date: | 2001-06-09 | | Release date: | 2001-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
Science, 294, 2001
|
|
1W3X
 
 | | Isopenicillin N synthase d-(L-a-aminoadipoyl)-(3R)-methyl-L-cysteine D-a-hydroxyisovaleryl ester complex (Oxygen exposed 5 minutes 20 bar) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N~6~-[(1R)-1-({[(1R,2R)-1-CARBOXY-3-HYDROXY-2-METHYLPROPYL]OXY}CARBONYL)-2-MERCAPTOPROP-2-EN-1-YL]-6-OXO-L-LYSINE | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2004-07-20 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Unexpected Oxidation of a Depsipeptide Substrate Analogue in Crystalline Isopenicillin N Synthase.
Chembiochem, 7, 2006
|
|
2CLK
 
 | | Tryptophan Synthase in complex with D-glyceraldehyde 3-phosphate (G3P) | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Ngo, H, Harris, R, Kimmich, N, Casino, P, Niks, D, Blumenstein, L, Barends, T.R, Kulik, V, Weyand, M, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-04-27 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Characterization of Allosteric Probes of Substrate Channeling in the Tryptophan Synthase Bienzyme Complex.
Biochemistry, 46, 2007
|
|
5UFF
 
 | | Crystal Structure of Variable Lymphocyte Receptor (VLR) RBC36 with Fucose(alpha-1-2)Lactose bound | | Descriptor: | RBC36, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Collins, B.C, Gunn, R.J, McKitrick, T.R, Herrin, B.R, Cummings, R.D, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2017-01-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Structural Insights into VLR Fine Specificity for Blood Group Carbohydrates.
Structure, 25, 2017
|
|
3UPL
 
 | | Crystal structure of the Brucella abortus enzyme catalyzing the first committed step of the methylerythritol 4-phosphate pathway. | | Descriptor: | GLYCEROL, MAGNESIUM ION, Oxidoreductase | | Authors: | Calisto, B.M, Perez-Gil, J, Fita, I, Rodriguez-Concepcion, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Brucella abortus deoxyxylulose-5-phosphate reductoisomerase-like (DRL) enzyme involved in isoprenoid biosynthesis.
J.Biol.Chem., 287, 2012
|
|
6D94
 
 | | Crystal structure of PPAR gamma in complex with Mediator of RNA polymerase II transcription subunit 1 | | Descriptor: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Mou, T.C, Chrisman, I.M, Hughes, T.S, Sprang, S.R. | | Deposit date: | 2018-04-27 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural mechanism of nuclear receptor biased agonism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3SRD
 
 | | Human M2 pyruvate kinase in complex with fructose 1-6 bisphosphate and Oxalate. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Morgan, H.P, O'Reilly, F, Palmer, R, McNae, I.W, Nowicki, M.W, Wear, M.A, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Allosetric regulation of M2 pyruvate kinase.
To be Published
|
|
3KAP
 
 | | Flavodoxin from Desulfovibrio desulfuricans ATCC 27774 (oxidized form) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Romero, A, Caldeira, J, LeGall, J, Moura, I, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2009-10-19 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of flavodoxin from Desulfovibrio desulfuricans ATCC 27774 in two oxidation states.
Eur.J.Biochem., 239, 1996
|
|
3J1Z
 
 | | Inward-Facing Conformation of the Zinc Transporter YiiP revealed by Cryo-electron Microscopy | | Descriptor: | Cation efflux family protein | | Authors: | Coudray, N, Valvo, S, Hu, M, Lasala, R, Kim, C, Vink, M, Zhou, M, Provasi, D, Filizola, M, Tao, J, Fang, J, Penczek, P.A, Ubarretxena-Belandia, I, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2012-07-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6DBA
 
 | | Crystal Structure of VHH R303 | | Descriptor: | nanobody VHH R303 | | Authors: | Brooks, C.L, Toride King, M, Huh, I. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of VHH-mediated neutralization of the food-borne pathogenListeria monocytogenes.
J. Biol. Chem., 293, 2018
|
|
1JT5
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag AND LEU 73 REPLACED BY VAL AND VAL 109 REPLACED BY LEU (L73V/V109L) | | Descriptor: | SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
1UO9
 
 | | Deacetoxycephalosporin C synthase complexed with succinate | | Descriptor: | DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (II) ION, SUCCINIC ACID | | Authors: | Valegard, K, Terwisscha Van scheltinga, A.C, Dubus, A, Oster, L.M, Rhangino, G, Hajdu, J, Andersson, I. | | Deposit date: | 2003-09-16 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structural Basis of Cephalosporin Formation in a Mononuclear Ferrous Enzyme
Nat.Struct.Mol.Biol., 11, 2004
|
|
6D7P
 
 | | Crystal Structure of Rat TRPV6*-Y466A | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Singh, A.K, Saotome, K, McGoldrick, L.L, Sobolevsky, A.I. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB.
Nat Commun, 9, 2018
|
|
6D7Q
 
 | | Crystal Structure of Rat TRPV6*-Y466A in complex with 2-Aminoethoxydiphenyl borate (2-APB) | | Descriptor: | 2-aminoethyl diphenylborinate, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, ... | | Authors: | Singh, A.K, Saotome, K, McGoldrick, L.L, Sobolevsky, A.I. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.497 Å) | | Cite: | Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB.
Nat Commun, 9, 2018
|
|
6D7X
 
 | | Crystal Structure of Rat TRPV6*-Y466A- in complex with brominated 2-Aminoethoxydiphenyl borate (2-APB-Br) | | Descriptor: | 2-aminoethyl (4-bromophenyl)phenylborinate, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, ... | | Authors: | Singh, A.K, Saotome, K, McGoldrick, L.L, Sobolevsky, A.I. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB.
Nat Commun, 9, 2018
|
|
2OSZ
 
 | |
1JHY
 
 | | Three-dimensional Structure of CobT in Complex with Phenol and Nicotinate | | Descriptor: | NICOTINIC ACID, Nicotinate Mononucleotide:5,6-Dimethylbenzimidazole Phosphoribosyltransferase, PHENOL, ... | | Authors: | Cheong, C.G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2001-06-28 | | Release date: | 2001-10-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of the biosynthesis of alternative lower ligands for cobamides by nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase from Salmonella enterica.
J.Biol.Chem., 276, 2001
|
|
