8BYH
 
 | |
6IKT
 
 | |
6IL0
 
 | |
6IL9
 
 | | One Glycerol complexed Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi | | Descriptor: | Fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi in complex with 1 glycerol, GLYCEROL, ZINC ION | | Authors: | Choi, M.Y, Kang, L.W, Ho, T.H, Nguyen, D.Q, Lee, I.H, Lee, J.H, Park, Y.S, Park, H.J. | | Deposit date: | 2018-10-17 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72005355 Å) | | Cite: | Crystal structure of fructuronate-tagaturonate epimerase UxaE from Cohnella laeviribosi
To Be Published
|
|
3ARV
 
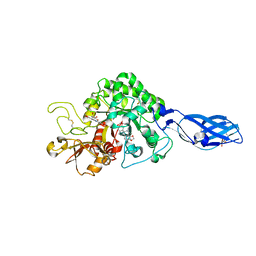 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with Sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3H3N
 
 | | Glycerol Kinase H232R with Glycerol | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glycerol kinase, ... | | Authors: | Yeh, J.I, Kettering, R.D. | | Deposit date: | 2009-04-16 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural characterizations of glycerol kinase: unraveling phosphorylation-induced long-range activation
Biochemistry, 48, 2009
|
|
3H46
 
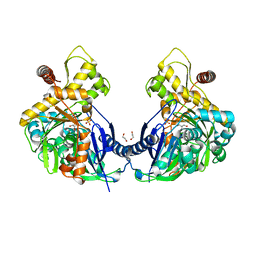 | | Glycerol Kinase H232E with Glycerol | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glycerol kinase, ... | | Authors: | Yeh, J.I, Kettering, R.D. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural characterizations of glycerol kinase: unraveling phosphorylation-induced long-range activation
Biochemistry, 48, 2009
|
|
8C1A
 
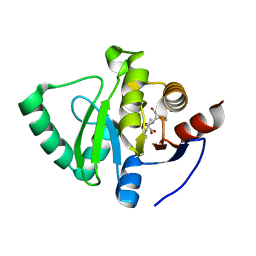 | |
3GJS
 
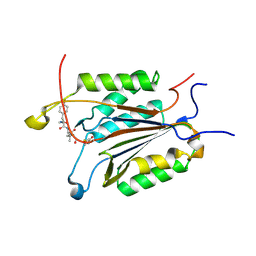 | | Caspase-3 Binds Diverse P4 Residues in Peptides | | Descriptor: | Ac-YVAD-Cho inhibitor, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Fang, B, Fu, G, Agniswamy, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2009-03-09 | | Release date: | 2009-03-24 | | Last modified: | 2011-08-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Caspase-3 binds diverse P4 residues in peptides as revealed by crystallography and structural modeling.
Apoptosis, 14, 2009
|
|
3H63
 
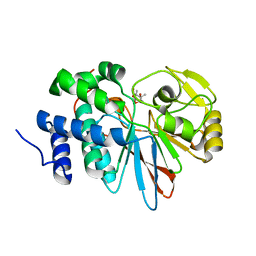 | | Catalytic domain of human Serine/Threonine Phosphatase 5 (PP5c) with two Mn2+ atoms originally soaked with cantharidin (which is present in the structure in the hydrolyzed form) | | Descriptor: | (1R,2S,3R,4S)-2,3-dimethyl-7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid, MANGANESE (II) ION, Serine/threonine-protein phosphatase 5 | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Talluri, E. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of serine/threonine phosphatase inhibition by the archetypal small molecules cantharidin and norcantharidin
J.Med.Chem., 52, 2009
|
|
5LYE
 
 | |
4L99
 
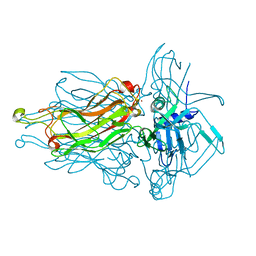 | | Structure of the RBP from lactococcal phage 1358 in complex with glycerol | | Descriptor: | GLYCEROL, Receptor Binding Protein, ZINC ION | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
1UCG
 
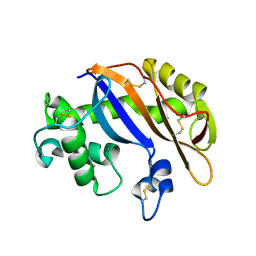 | | Crystal structure of Ribonuclease MC1 N71T mutant | | Descriptor: | MANGANESE (II) ION, Ribonuclease MC | | Authors: | Suzuki, A, Numata, T, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2003-04-14 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the ribonuclease MC1 mutants N71T and N71S in complex with 5'-GMP: structural basis for alterations in substrate specificity
Biochemistry, 42, 2003
|
|
5V0Y
 
 | | Solution structure of arenicin-3. | | Descriptor: | arenicin-3 | | Authors: | Edwards, I.A, Mobli, M. | | Deposit date: | 2017-02-28 | | Release date: | 2018-08-08 | | Last modified: | 2020-06-10 | | Method: | SOLUTION NMR | | Cite: | An amphipathic peptide with antibiotic activity against multidrug-resistant Gram-negative bacteria
Nat Commun, 2020
|
|
5V2A
 
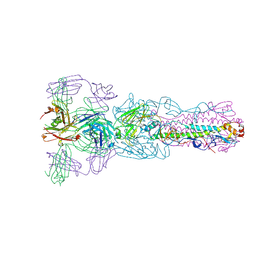 | | Crystal structure of Fab H7.167 in complex with influenza virus hemagglutinin from A/Shanghai/02/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H7.167 antibody, Hemagglutinin, ... | | Authors: | Zhang, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2017-03-03 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.656 Å) | | Cite: | H7N9 influenza virus neutralizing antibodies that possess few somatic mutations.
J. Clin. Invest., 126, 2016
|
|
7LXL
 
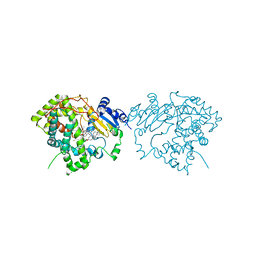 | | Crystal structure of human CYP3A4 bound to the testosterone dimer | | Descriptor: | 17alpha-hydroxy-7alpha-[(2Z)-4-(17beta-hydroxy-3-oxo-8alpha-androst-4-en-7beta-yl)but-2-en-1-yl]-8alpha,10alpha,13alpha,14beta-androst-4-en-3-one, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2021-03-04 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Innovative C 2 -symmetric testosterone and androstenedione dimers: Design, synthesis, biological evaluation on prostate cancer cell lines and binding study to recombinant CYP3A4.
Eur.J.Med.Chem., 220, 2021
|
|
6MAJ
 
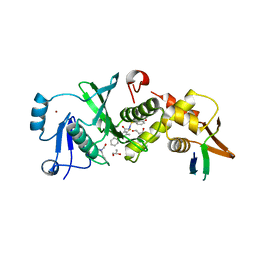 | | HBO1 is required for the maintenance of leukaemia stem cells | | Descriptor: | 4-fluoro-N'-[(3-hydroxyphenyl)sulfonyl]-5-methyl[1,1'-biphenyl]-3-carbohydrazide, BRD1 protein, GLYCEROL, ... | | Authors: | Ren, B, Peat, T.S, Monahan, B, Dawson, M, Street, I. | | Deposit date: | 2018-08-27 | | Release date: | 2019-12-25 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.139 Å) | | Cite: | HBO1 is required for the maintenance of leukaemia stem cells.
Nature, 577, 2020
|
|
1UE4
 
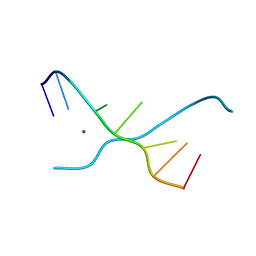 | | Crystal structure of d(GCGAAAGC) | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*AP*GP*C)-3', MAGNESIUM ION | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G.A pairs to form a functional G.AxA.G crossing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UDU
 
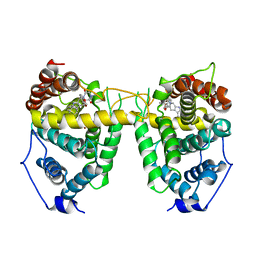 | | Crystal structure of Human Phosphodiesterase 5 complexed with tadalafil(Cialis) | | Descriptor: | 6-BENZO[1,3]DIOXOL-5-YL-2-METHYL-2,3,6,7,12,12A-HEXAHYDRO-PYRAZINO[1',2':1,6]PYRIDO[3,4-B]INDOLE-1,4-DIONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Sung, B.-J, Lee, J.I, Heo, Y.-S, Kim, J.H, Moon, J, Yoon, J.M, Hyun, Y.-L, Kim, E, Eum, S.J, Lee, T.G, Cho, J.M, Park, S.-Y, Lee, J.-O, Jeon, Y.H, Hwang, K.Y, Ro, S. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules
Nature, 425, 2003
|
|
6A28
 
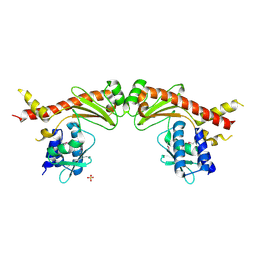 | | Crystal structure of PprA W183R mutant form 2 | | Descriptor: | DNA repair protein PprA, SULFATE ION | | Authors: | Adachi, M, Shibazaki, C, Shimizu, R, Arai, S, Satoh, K, Narumi, I, Kuroki, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Extended structure of pleiotropic DNA repair-promoting protein PprA from Deinococcus radiodurans.
FASEB J., 33, 2019
|
|
7NH4
 
 | |
4JAV
 
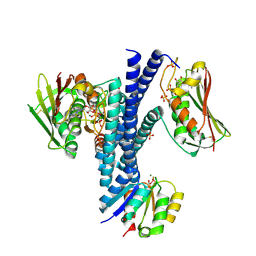 | | Structural basis of a rationally rewired protein-protein interface (HK853wt and RR468mutant V13P, L14I, I17M and N21V) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Histidine kinase, ... | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
7NH5
 
 | |
3AS0
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with Sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
1YH3
 
 | | Crystal structure of human CD38 extracellular domain | | Descriptor: | ADP-ribosyl cyclase 1 | | Authors: | Liu, Q, Kriksunov, I.A, Graeff, R, Munshi, C, Lee, H.C, Hao, Q. | | Deposit date: | 2005-01-06 | | Release date: | 2005-09-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of human CD38 extracellular domain.
Structure, 13, 2005
|
|
