9B67
 
 | |
9CD5
 
 | | FGFR1 Kinase Domain Soak with Inhibitor TYRA-300 | | Descriptor: | (3P)-5-[(1R)-1-(3,5-dichloropyridin-4-yl)ethoxy]-3-{6-[6-(methanesulfonyl)-2,6-diazaspiro[3.3]heptan-2-yl]pyridin-3-yl}-1H-indazole, 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, ... | | Authors: | Hoffman, I.H, Nelson, K.J, Rideout, M.C, Frye, C, Bensen, D.C, Hudkins, R.L. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.939 Å) | | Cite: | Discovery of TYRA-300: First Oral Selective FGFR3 Inhibitor for the Treatment of Urothelial Cancers and Achondroplasia.
J.Med.Chem., 67, 2024
|
|
9PAP
 
 | |
9FT1
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 13 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9FT0
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 42 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9MSI
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18N | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
9C1B
 
 | | Crystal structure of GDP-bound human M-RAS protein in crystal form II | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bester, S.M, Abrahamsen, R, Samora, L.R, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-05-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of the GDP-bound human M-RAS protein in two crystal forms.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9C1A
 
 | | Crystal structure of GDP-bound human M-RAS protein in crystal form I | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bester, S.M, Abrahamsen, R, Samora, L.R, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-05-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of the GDP-bound human M-RAS protein in two crystal forms.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
9B00
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with berberine analog of chloramphenicol CAM-BER, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.80A resolution | | Descriptor: | 13-(2-{[(1R,2R)-1,3-dihydroxy-1-(4-nitrophenyl)propan-2-yl]amino}-2-oxoethyl)-9,10-dimethoxy-5,6-dihydro-2H-[1,3]dioxolo[4,5-g]isoquinolino[3,2-a]isoquinolin-7-ium, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Batool, Z, Pavlova, J.A, Paranjpe, M.N, Tereshchenkov, A.G, Lukianov, D.A, Osterman, I.A, Bogdanov, A.A, Sumbatyan, N.V, Polikanov, Y.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Berberine analog of chloramphenicol exhibits a distinct mode of action and unveils ribosome plasticity.
Structure, 32, 2024
|
|
9FSV
 
 | | Yeast 20S proteasome with human beta2i (1-53) in complex with epoxyketone inhibitor 42 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-22 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9CD7
 
 | | FGFR3 Kinase Domain with Inhibitor TYRA-300 | | Descriptor: | (3P)-5-[(1R)-1-(3,5-dichloropyridin-4-yl)ethoxy]-3-{6-[6-(methanesulfonyl)-2,6-diazaspiro[3.3]heptan-2-yl]pyridin-3-yl}-1H-indazole, Fibroblast growth factor receptor 3, GLYCEROL, ... | | Authors: | Hoffman, I.H, Nelson, K.J, Rideout, M.C, Nix, J.C, Frye, C, Bensen, D.C, Hudkins, R.L. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of TYRA-300: First Oral Selective FGFR3 Inhibitor for the Treatment of Urothelial Cancers and Achondroplasia.
J.Med.Chem., 67, 2024
|
|
6ZXS
 
 | | Cold grown Pea Photosystem I | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Caspy, I, Borovikova-Sheinker, A, Subramanyam, R, Nelson, N. | | Deposit date: | 2020-07-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of cold grown pea Photosystem I
To Be Published
|
|
7QNG
 
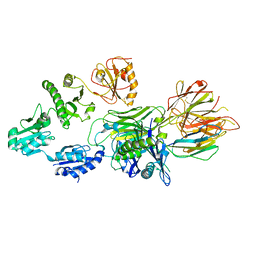 | | Structure of a MHC I-Tapasin-ERp57 complex | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Mueller, I.K, Thomas, C, Trowitzsch, S, Tampe, R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an MHC I-tapasin-ERp57 editing complex defines chaperone promiscuity.
Nat Commun, 13, 2022
|
|
6WYV
 
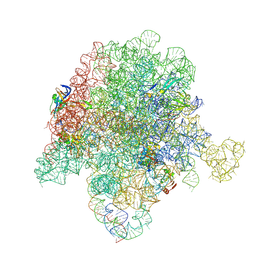 | | E. coli 50S ribosome bound to compounds 47 and VS1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6UF4
 
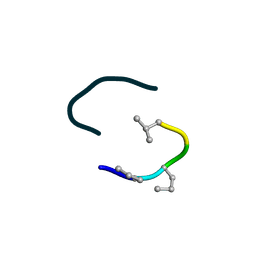 | | S2 symmetric peptide design number 4 crystal form 2, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 2 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-23 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UF9
 
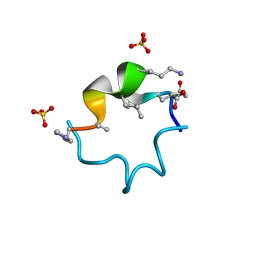 | | S4 symmetric peptide design number 1, Tim apo form | | Descriptor: | S4-1, Tim apo-form, SULFATE ION | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-24 | | Release date: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6U5E
 
 | | RT XFEL structure of CypA solved using celloluse carrier media | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Wolff, A.M, Nango, E, Nakane, T, Young, I.D, Brewster, A.S, Sugahara, M, Tanaka, R, Sauter, N.K, Tono, K, Iwata, S, Fraser, J.S, Thompson, M.C. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
Iucrj, 7, 2020
|
|
6TXW
 
 | | V30G Transthyretin structure in complex with Tolcalpone | | Descriptor: | Tolcapone, Transthyretin | | Authors: | Varejao, N, Reverter, D, Pinheiro, F, Pallares, I, Ventura, S. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.153 Å) | | Cite: | Tolcapone, a potent aggregation inhibitor for the treatment of familial leptomeningeal amyloidosis.
Febs J., 288, 2021
|
|
6KLK
 
 | |
6KEI
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 16-methoxy-11-methyl-6-[(pyridin-2-yl)methoxy]-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one | | Descriptor: | 16-methoxy-11-methyl-6-[(pyridin-2-yl)methoxy]-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
6D7Q
 
 | | Crystal Structure of Rat TRPV6*-Y466A in complex with 2-Aminoethoxydiphenyl borate (2-APB) | | Descriptor: | 2-aminoethyl diphenylborinate, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, ... | | Authors: | Singh, A.K, Saotome, K, McGoldrick, L.L, Sobolevsky, A.I. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.497 Å) | | Cite: | Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB.
Nat Commun, 9, 2018
|
|
6D7X
 
 | | Crystal Structure of Rat TRPV6*-Y466A- in complex with brominated 2-Aminoethoxydiphenyl borate (2-APB-Br) | | Descriptor: | 2-aminoethyl (4-bromophenyl)phenylborinate, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, ... | | Authors: | Singh, A.K, Saotome, K, McGoldrick, L.L, Sobolevsky, A.I. | | Deposit date: | 2018-04-25 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB.
Nat Commun, 9, 2018
|
|
6KIO
 
 | | Complex of yeast cytoplasmic dynein MTBD-High and MT without DTT | | Descriptor: | Dynein heavy chain, cytoplasmic, Tubulin alpha-1A chain, ... | | Authors: | Komori, Y, Nishida, N, Shimada, I, Kikkawa, M. | | Deposit date: | 2019-07-19 | | Release date: | 2020-03-04 | | Last modified: | 2020-03-11 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Structural basis for two-way communication between dynein and microtubules.
Nat Commun, 11, 2020
|
|
6PCR
 
 | | E. coli 50S ribosome bound to compound 40o | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,26aR)-14-hydroxy-4,12-dimethyl-1,7,16,22-tetraoxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl (2-bromopyridin-4-yl)carbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-06-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6U7L
 
 | | 2.75 Angstrom Crystal Structure of Galactarate Dehydratase from Escherichia coli. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Galactarate dehydratase (L-threo-forming) | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-06 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of galactarate dehydratase, a new fold in an enolase involved in bacterial fitness after antibiotic treatment.
Protein Sci., 29, 2020
|
|
