3CR8
 
 | | Hexameric APS kinase from Thiobacillus denitrificans | | Descriptor: | Sulfate adenylyltransferase, adenylylsulfate kinase | | Authors: | Gay, S.C, Segel, I.H, Fisher, A.J. | | Deposit date: | 2008-04-04 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the two-domain hexameric APS kinase from Thiobacillus denitrificans: structural basis for the absence of ATP sulfurylase activity.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3CVW
 
 | | Drosophila melanogaster (6-4) photolyase H365N mutant bound to ds DNA with a T-T (6-4) photolesion and cofactor F0 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a cofactor f0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
2YC0
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH R-2-HYDROXYGLUTARATE | | Descriptor: | (2R)-2-hydroxypentanedioic acid, FE (II) ION, GLYCEROL, ... | | Authors: | Chowdhury, R, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2011-03-10 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
EMBO Rep., 12, 2011
|
|
5YP3
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) with Ile-Pro from Pseudoxanthomonas mexicana | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL, ISOLEUCINE, ... | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
4R5G
 
 | | Crystal structure of the DnaK C-terminus with the inhibitor PET-16 | | Descriptor: | Chaperone protein DnaK, triphenyl(phenylethynyl)phosphonium | | Authors: | Leu, J.I, Zhang, P, Murphy, M.E, Marmorstein, R, George, D.L. | | Deposit date: | 2014-08-21 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4501 Å) | | Cite: | Structural Basis for the Inhibition of HSP70 and DnaK Chaperones by Small-Molecule Targeting of a C-Terminal Allosteric Pocket.
Acs Chem.Biol., 9, 2014
|
|
2Y6A
 
 | | Ascorbate Peroxidase R38A mutant | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Metcalfe, C.L, Efimov, I, Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-01-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proton Delivery to Ferryl Heme in a Heme Peroxidase: Enzymatic Use of the Grotthuss Mechanism.
J.Am.Chem.Soc., 133, 2011
|
|
3GD3
 
 | |
2HMP
 
 | | Uncomplexed actin cleaved with protease ECP32 | | Descriptor: | 1,2-ETHANEDIOL, 2,2',2''-NITRILOTRIETHANOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Klenchin, V.A, Khaitlina, S.Y, Rayment, I. | | Deposit date: | 2006-07-11 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Crystal structure of polymerization-competent actin
J.Mol.Biol., 362, 2006
|
|
4R0W
 
 | |
4RGT
 
 | | 2.0 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with 3-N-Acetylneuraminyl-N-acetyllactosamine. | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Putative uncharacterized protein | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Filippova, E.V, Halavaty, A, Dubrovska, I, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus in Complex with 3-N-Acetylneuraminyl-N-acetyllactosamine.
TO BE PUBLISHED
|
|
3P66
 
 | | Time-dependent and Protein-directed In Situ Growth of Gold Nanoparticles in a Single Crystal of Lysozyme | | Descriptor: | GOLD 3+ ION, Lysozyme C | | Authors: | Wei, H, Wang, Z, Zhang, J, House, S, Gao, Y.-G, Yang, L, Robinson, H, Tan, L.H, Xing, H, Hou, C, Robertson, I.M, Zuo, J.-M, Lu, Y. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme.
Nat Nanotechnol, 6, 2011
|
|
2YDE
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH S-2-HYDROXYGLUTARATE | | Descriptor: | (2S)-2-HYDROXYPENTANEDIOIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Chowdhury, R, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2011-03-18 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
EMBO Rep., 12, 2011
|
|
6K1G
 
 | | Crystal structure of the L-fucose isomerase soaked with Mn2+ from Raoultella sp. | | Descriptor: | L-fucose isomerase, MANGANESE (II) ION | | Authors: | Kim, I.J, Kim, D.H, Nam, K.H, Kim, K.H. | | Deposit date: | 2019-05-10 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Enzymatic synthesis of l-fucose from l-fuculose using a fucose isomerase fromRaoultellasp. and the biochemical and structural analyses of the enzyme.
Biotechnol Biofuels, 12, 2019
|
|
7LOP
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CV05-163 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab heavy chain, CR3022 Fab light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-02-10 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants.
Science, 373, 2021
|
|
1TKK
 
 | | The Structure of a Substrate-Liganded Complex of the L-Ala-D/L-Glu Epimerase from Bacillus subtilis | | Descriptor: | ALANINE, GLUTAMIC ACID, MAGNESIUM ION, ... | | Authors: | Klenchin, V.A, Schmidt, D.M, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of Enzymatic Activities in the Enolase Superfamily: Structure of a Substrate-Liganded Complex of the l-Ala-d/l-Glu Epimerase from Bacillus subtilis(,).
Biochemistry, 43, 2004
|
|
3CVX
 
 | | Drosophila melanogaster (6-4) photolyase H369M mutant bound to ds DNA with a T-T (6-4) photolesion | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a coenzyme F0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
3CN4
 
 | |
2LO8
 
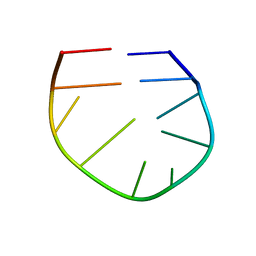 | |
3G51
 
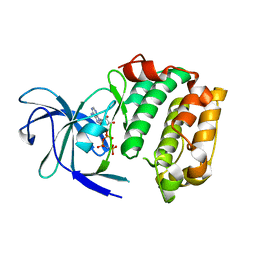 | |
4RGZ
 
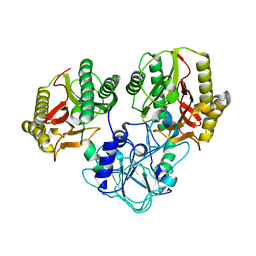 | | Crystal structure of recombinant prolidase from Thermococcus sibiricus at P21221 spacegroup | | Descriptor: | PHOSPHATE ION, Xaa-Pro aminopeptidase, ZINC ION | | Authors: | Timofeev, V.I, Korgenevsky, D.A, Gorbacheva, M.A, Boyko, K.M, Slutsky, E, Rakitina, T.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2014-10-01 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of recombinant prolidase from Thermococcus sibiricus in space group P21221.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2HG5
 
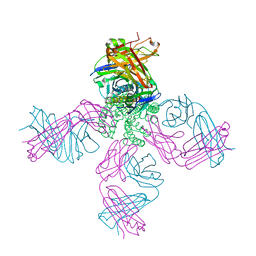 | | Cs+ complex of a K channel with an amide to ester substitution in the selectivity filter | | Descriptor: | (2S)-2-(BUTYRYLOXY)-3-HYDROXYPROPYL NONANOATE, CESIUM ION, FAB HEAVY CHAIN, ... | | Authors: | Valiyaveetil, F.I, MacKinnon, R, Muir, T.W. | | Deposit date: | 2006-06-26 | | Release date: | 2006-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Functional Consequences of an Amide-to-Ester Substitution in the Selectivity Filter of a Potassium Channel.
J.Am.Chem.Soc., 128, 2006
|
|
2LO5
 
 | | Structural Basis for Bifunctional Zn(II) Macrocyclic Complex Recognition of Thymine Bulges in DNA, Structure of a Thymine bulge | | Descriptor: | DNA (5'-D(*GP*GP*CP*CP*GP*CP*AP*GP*TP*GP*CP*C)-3') | | Authors: | Morrow, J.R, Fountain, M.A, del Mundo, I.A, Siter, K.E. | | Deposit date: | 2012-01-17 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Bifunctional Zinc(II) Macrocyclic Complex Recognition of Thymine Bulges in DNA.
Inorg.Chem., 51, 2012
|
|
8UYS
 
 | | SARS-CoV-2 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-2 RNA SL5 domain. | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3CZL
 
 | |
8UYJ
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
