7AE8
 
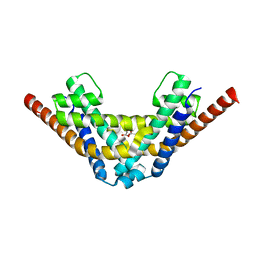 | | Crystal structure of HEPN(R102A) toxin | | Descriptor: | ACETATE ION, HEPN toxin | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
6ZXG
 
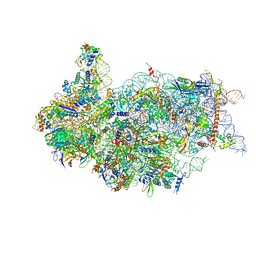 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State H1 | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
6ZTY
 
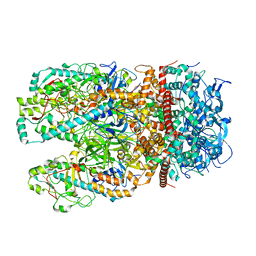 | |
4URT
 
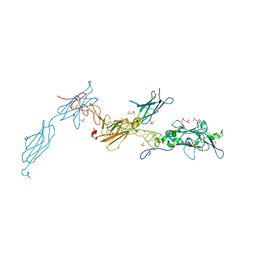 | | The crystal structure of a fragment of netrin-1 in complex with FN5- FN6 of DCC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Finci, L.I, Krueger, N, Sun, X, Zhang, J, Chegkazi, M, Wu, Y, Schenk, G, Mertens, H.D.T, Svergun, D.I, Zhang, Y, Wang, J.-h, Meijers, R. | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Crystal Structure of Netrin-1 in Complex with Dcc Reveals the Bi-Functionality of Netrin-1 as a Guidance Cue
Neuron, 83, 2014
|
|
4WXG
 
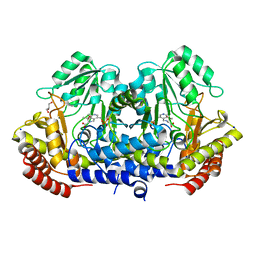 | | Crystal structure of L-Serine Hydroxymethyltransferase in complex with a mixture of L-Threonine and Glycine | | Descriptor: | GLYCEROL, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-threonine, SODIUM ION, ... | | Authors: | Hernandez, K, Zelen, I, Petrillo, G, Uson, I, Wandtke, C, Bujons, J, Joglar, J, Parella, T, Clapes, P. | | Deposit date: | 2014-11-13 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered L-Serine Hydroxymethyltransferase from Streptococcus thermophilus for the Synthesis of alpha , alpha-Dialkyl-alpha-Amino Acids.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1LW2
 
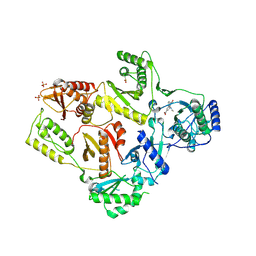 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 1051U91 | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
4WWK
 
 | | Crystal structure of human TCR Alpha Chain-TRAV12-3, Beta Chain-TRBV6-5, Antigen-presenting molecule CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, A, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
1LW0
 
 | | CRYSTAL STRUCTURE OF T215Y MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-30 | | Release date: | 2002-10-30 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
6OCR
 
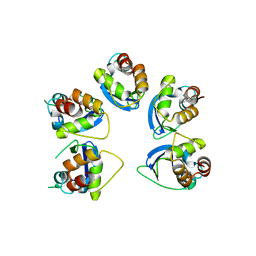 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1MHE
 
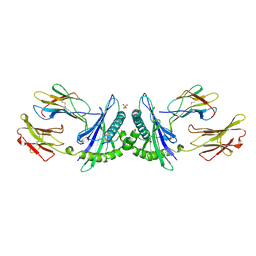 | | THE HUMAN NON-CLASSICAL MAJOR HISTOCOMPATIBILITY COMPLEX MOLECULE HLA-E | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN HLA-E, PEPTIDE (VMAPRTVLL), ... | | Authors: | O'Callaghan, C.A, Tormo, J, Willcox, B.E, Braud, V.B, Jakobsen, B.K, Stuart, D.I, Mcmichael, A.J, Bell, J.I, Jones, E.Y. | | Deposit date: | 1998-08-24 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E.
Mol.Cell, 1, 1998
|
|
4UXS
 
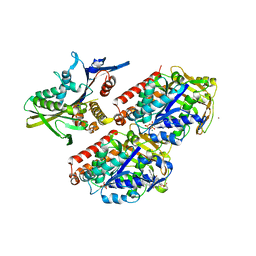 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN-3 MOTOR DOMAIN, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
6OF1
 
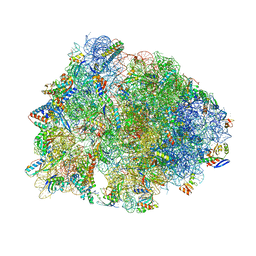 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with dirithromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khabibullina, N.F, Tereshchenkov, A.G, Komarova, E.S, Syroegin, E.A, Shiriaev, D.I, Paleskava, A, Kartsev, V.G, Bogdanov, A.A, Konevega, A.L, Dontsova, O.A, Sergiev, P.V, Osterman, I.A, Polikanov, Y.S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Dirithromycin Bound to the Bacterial Ribosome Suggests New Ways for Rational Improvement of Macrolides.
Antimicrob.Agents Chemother., 63, 2019
|
|
1WAY
 
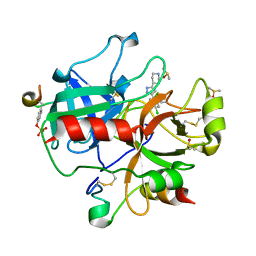 | | Active site thrombin inhibitors | | Descriptor: | 4-[3-(4-CHLOROPHENYL)-1H-PYRAZOL-5-YL]PIPERIDINE, DIMETHYL SULFOXIDE, HIRUGEN, ... | | Authors: | Hartshorn, M.J, Murray, C.W, Cleasby, A, Frederickson, M, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-10-28 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
1WBG
 
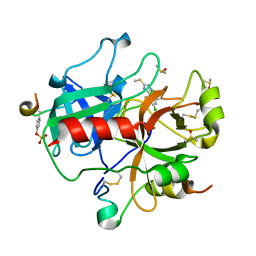 | | Active site thrombin inhibitors | | Descriptor: | 3-(4-CHLOROPHENYL)-5-(METHYLTHIO)-4H-1,2,4-TRIAZOLE, DIMETHYL SULFOXIDE, HIRUGEN, ... | | Authors: | Hartshorn, M.J, Murray, C.W, Cleasby, A, Frederickson, M, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-11-01 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
4WX1
 
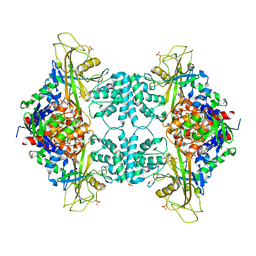 | |
4X35
 
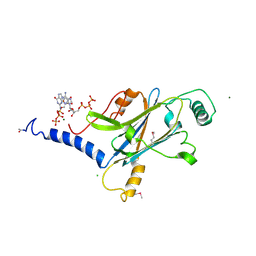 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | Deposit date: | 2014-11-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
4X6G
 
 | |
6ZQP
 
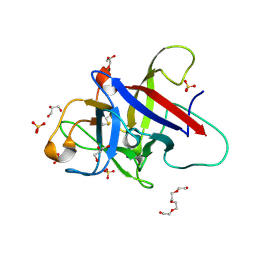 | | Structure of the Pmt2-MIR domain with bound ligands | | Descriptor: | GLYCEROL, PMT2 isoform 1, SULFATE ION, ... | | Authors: | Wild, K, Chiapparino, A, Hackmann, Y, Mortensen, S, Sinning, I. | | Deposit date: | 2020-07-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional implications of MIR domains in protein O -mannosylation.
Elife, 9, 2020
|
|
6ZQV
 
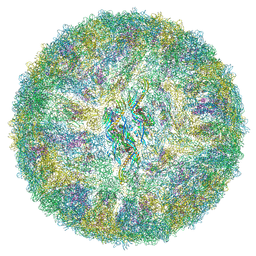 | | Cryo-EM structure of mature Spondweni virus | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
6ZRC
 
 | |
7A5V
 
 | | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-22 | | Release date: | 2020-11-18 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
6ZMP
 
 | |
4UV3
 
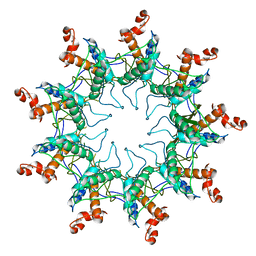 | | Structure of the curli transport lipoprotein CsgG in its membrane- bound conformation | | Descriptor: | CURLI PRODUCTION ASSEMBLY/TRANSPORT COMPONENT CSGG | | Authors: | Goyal, P, Krasteva, P.V, Gerven, N.V, Gubellini, F, Broeck, I.V.D, Troupiotis-Tsailaki, A, Jonckheere, W, Pehau-Arnaudet, G, Pinkner, J.S, Chapman, M.R, Hultgren, S.J, Howorka, S, Fronzes, R, Remaut, H. | | Deposit date: | 2014-08-04 | | Release date: | 2014-09-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural and Mechanistic Insights Into the Bacterial Amyloid Secretion Channel Csgg.
Nature, 516, 2014
|
|
4V1L
 
 | | High resolution structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
