1GSF
 
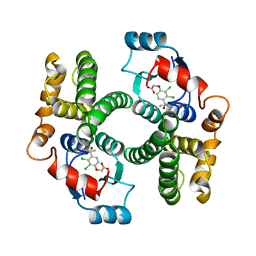 | | GLUTATHIONE TRANSFERASE A1-1 COMPLEXED WITH ETHACRYNIC ACID | | Descriptor: | ETHACRYNIC ACID, GLUTATHIONE TRANSFERASE A1-1 | | Authors: | L'Hermite, G, Sinning, I, Cameron, A.D, Jones, T.A. | | Deposit date: | 1995-06-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of human alpha-class glutathione transferase A1-1 in the apo-form and in complexes with ethacrynic acid and its glutathione conjugate.
Structure, 3, 1995
|
|
1HHS
 
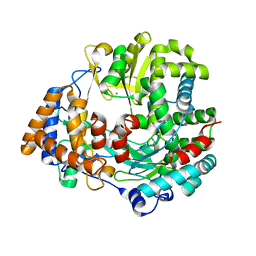 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2000-12-28 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
6ZMK
 
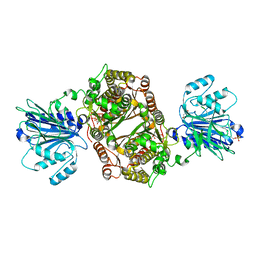 | | Crystal structure of human GFAT-1 L405R | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Mayr, F, Miethe, S, Atanassov, I, Baumann, U, Denzel, M.S. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1.
Nat Commun, 12, 2021
|
|
6KX1
 
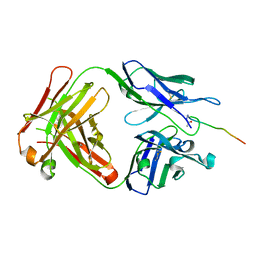 | | Crystal structure of SN-101 mAb in complex with MUC1 glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain, ... | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
1HHT
 
 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 plus template | | Descriptor: | DNA (5'-(*TP*TP*TP*CP*C)-3'), MANGANESE (II) ION, P2 PROTEIN | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2000-12-28 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
5O0Z
 
 | | Structure of laspartomycin C in complex with geranyl-phosphate | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vlieg, H.C, Kleijn, L.H.J, Martin, N.I, Janssen, B.J.C. | | Deposit date: | 2017-05-17 | | Release date: | 2017-11-15 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A High-Resolution Crystal Structure that Reveals Molecular Details of Target Recognition by the Calcium-Dependent Lipopeptide Antibiotic Laspartomycin C.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7RSI
 
 | | The cryo-EM map of KIF18A bound to KIFBP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KIF-binding protein, Kinesin-like protein KIF18A, ... | | Authors: | Tan, Z, Solon, A.L, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
6RC9
 
 | |
5O5C
 
 | | The crystal structure of DfoJ, the desferrioxamine biosynthetic pathway lysine decarboxylase from the fire blight disease pathogen Erwinia amylovora | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase involved in desferrioxamine biosynthesis | | Authors: | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
7RSQ
 
 | | Cryo-EM structure of KIFBP core | | Descriptor: | KIF-binding protein | | Authors: | Solon, A.L, Tan, Z, Schutt, K.L, Jepsen, L, Haynes, S.E, Nesvizhskii, A.I, Sept, D, Stumpff, J, Ohi, R, Cianfrocco, M.A. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Kinesin-binding protein remodels the kinesin motor to prevent microtubule binding.
Sci Adv, 7, 2021
|
|
1I74
 
 | | STREPTOCOCCUS MUTANS INORGANIC PYROPHOSPHATASE | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PROBABLE MANGANESE-DEPENDENT INORGANIC PYROPHOSPHATASE, ... | | Authors: | Merckel, M.C, Fabrichniy, I.P, Goldman, A, Lahti, R, Salminen, A. | | Deposit date: | 2001-03-07 | | Release date: | 2001-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Streptococcus mutans pyrophosphatase: a new fold for an old mechanism.
Structure, 9, 2001
|
|
6UNL
 
 | | CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, tert-butyl [(2R)-1-(naphthalen-1-yl)-3-{[(2S)-3-(naphthalen-1-yl)-1-oxo-1-{[(pyridin-3-yl)methyl]amino}propan-2-yl]sulfanyl}propan-2-yl]carbamate | | Authors: | Sevrioukova, I. | | Deposit date: | 2019-10-12 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An increase in side-group hydrophobicity largely improves the potency of ritonavir-like inhibitors of CYP3A4.
Bioorg.Med.Chem., 28, 2020
|
|
6RFB
 
 | | Crystal structure of the potassium-pumping S254A mutant of the light-driven sodium pump KR2 in the monomeric form, pH 4.3 | | Descriptor: | EICOSANE, GLYCEROL, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
2CBI
 
 | | Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase | | Descriptor: | CHLORIDE ION, GAMMA-BUTYROLACTONE, GLYCEROL, ... | | Authors: | Rao, F.V, Dorfmueller, H.C, Villa, F, Allwood, M, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2006-01-04 | | Release date: | 2006-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis.
EMBO J., 25, 2006
|
|
6RF5
 
 | | Crystal structure of the light-driven sodium pump KR2 in the monomeric form, pH 6.0 | | Descriptor: | EICOSANE, GLYCEROL, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6RF6
 
 | | Crystal structure of the light-driven sodium pump KR2 in the monomeric form, pH 8.0 | | Descriptor: | EICOSANE, RETINAL, SODIUM ION, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
1ICF
 
 | | CRYSTAL STRUCTURE OF MHC CLASS II ASSOCIATED P41 II FRAGMENT IN COMPLEX WITH CATHEPSIN L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CATHEPSIN L: HEAVY CHAIN), PROTEIN (CATHEPSIN L: LIGHT CHAIN), ... | | Authors: | Guncar, G, Pungercic, G, Klemencic, I, Turk, V, Turk, D. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S.
EMBO J., 18, 1999
|
|
6RFA
 
 | | Crystal structure of the H30K mutant of the light-driven sodium pump KR2 in the monomeric form, pH 8.0 | | Descriptor: | EICOSANE, GLYCEROL, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
1RUI
 
 | | RHINOVIRUS 14 MUTANT S1223G COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52084 | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1RCO
 
 | | SPINACH RUBISCO IN COMPLEX WITH THE INHIBITOR D-XYLULOSE-2,2-DIOL-1,5-BISPHOSPHATE | | Descriptor: | D-XYLULOSE-2,2-DIOL-1,5-BISPHOSPHATE, RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE | | Authors: | Taylor, T.C, Andersson, I. | | Deposit date: | 1996-10-31 | | Release date: | 1997-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A common structural basis for the inhibition of ribulose 1,5-bisphosphate carboxylase by 4-carboxyarabinitol 1,5-bisphosphate and xylulose 1,5-bisphosphate.
J.Biol.Chem., 271, 1996
|
|
8OEI
 
 | | SFX structure of FutA after an accumulated dose of 350 kGy | | Descriptor: | FE (III) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7XBS
 
 | | Crystal structure of the adenylation domain of CmnG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CmnG | | Authors: | Chen, I.H, Wang, Y.L, Chang, C.Y. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization and Structural Determination of CmnG-A, the Adenylation Domain That Activates the Nonproteinogenic Amino Acid Capreomycidine in Capreomycin Biosynthesis.
Chembiochem, 23, 2022
|
|
6ZMJ
 
 | | Crystal structure of human GFAT-1 R203H | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Mayr, F, Miethe, S, Atanassov, I, Baumann, U, Denzel, M.S. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.774 Å) | | Cite: | Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1.
Nat Commun, 12, 2021
|
|
7D36
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(3S)-1-amino-5-fluoro-3-methyl-3,4-dihydro-2,6-naphthyridin-3-yl]-4-fluorophenyl}-5-cyano-3-methylpyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Nakahara, K, Mitsuoka, Y, Kasuya, S, Yamamoto, T, Yamamoto, S, Ito, H, Kido, Y, Kusakabe, K.I. | | Deposit date: | 2020-09-18 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Balancing potency and basicity by incorporating fluoropyridine moieties: Discovery of a 1-amino-3,4-dihydro-2,6-naphthyridine BACE1 inhibitor that affords robust and sustained central A beta reduction.
Eur.J.Med.Chem., 216, 2021
|
|
7XBU
 
 | | Crystal structure of the adenylation domain of CmnG in complex with capreomycidine | | Descriptor: | (2S)-amino[(4R)-2-amino-1,4,5,6-tetrahydropyrimidin-4-yl]ethanoic acid, CmnG | | Authors: | Chen, I.H, Wang, Y.L, Chang, C.Y. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization and Structural Determination of CmnG-A, the Adenylation Domain That Activates the Nonproteinogenic Amino Acid Capreomycidine in Capreomycin Biosynthesis.
Chembiochem, 23, 2022
|
|
