1SWR
 
 | | CORE-STREPTAVIDIN MUTANT W120A IN COMPLEX WITH BIOTIN AT PH 7.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
7QAD
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with 1,4-Oxazepane hydrochloride | | Descriptor: | 1,4-oxazepane, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors.
To Be Published
|
|
1SWP
 
 | | CORE-STREPTAVIDIN MUTANT W120F IN COMPLEX WITH BIOTIN AT PH 7.5 | | Descriptor: | BIOTIN, CORE-STREPTAVIDIN, EPI-BIOTIN | | Authors: | Freitag, S, Le Trong, I, Chilkoti, A, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-01-27 | | Release date: | 1999-02-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of binding site tryptophan mutants in the high-affinity streptavidin-biotin complex.
J.Mol.Biol., 279, 1998
|
|
7QD3
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with morpholine | | Descriptor: | Cholinephosphate cytidylyltransferase, morpholine | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-11-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
6DTD
 
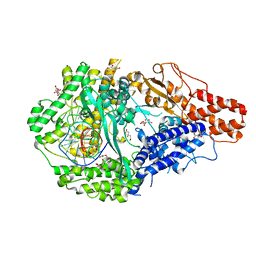 | |
5NZJ
 
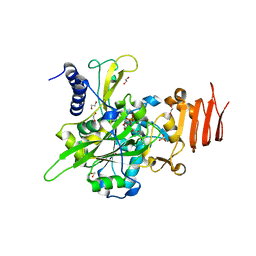 | | Crystal structure of UDP-glucose pyrophosphorylase G45Y mutant from Leishmania major in complex with UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UDP-glucose pyrophosphorylase, ... | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
6KX0
 
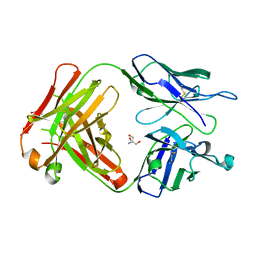 | | Crystal structure of SN-101 mAb non-liganded form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
1M7L
 
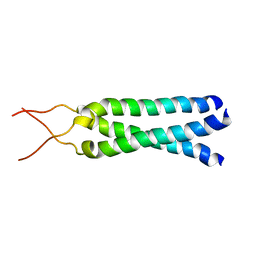 | | Solution Structure of the Coiled-Coil Trimerization Domain from Lung Surfactant Protein D | | Descriptor: | Pulmonary surfactant-associated protein D | | Authors: | Kovacs, H, O'Donoghue, S.I, Hoppe, H.-J, Comfort, D, Reid, K.B.M, Campbell, I.D, Nilges, M. | | Deposit date: | 2002-07-22 | | Release date: | 2002-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the coiled-coil trimerization domain from lung surfactant protein D
J.BIOMOL.NMR, 24, 2002
|
|
2BR7
 
 | | Crystal Structure of Acetylcholine-binding Protein (AChBP) from Aplysia californica in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Celie, P.H.N, Kasheverov, I.E, Mordvintsev, D.Y, Hogg, R.C, Van Nierop, P, Van Elk, R, Van Rossum-Fikkert, S.E, Zhmak, M.N, Bertrand, D, Tsetlin, V, Sixma, T.K, Smit, A.B. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Nicotinic Acetylcholine Receptor Homolog Achbp in Complex with an Alpha- Conotoxin Pnia Variant
Nat.Struct.Mol.Biol., 12, 2005
|
|
4CAT
 
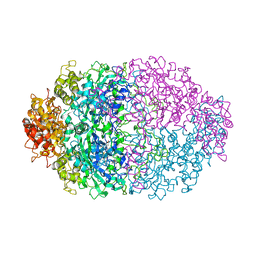 | | THREE-DIMENSIONAL STRUCTURE OF CATALASE FROM PENICILLIUM VITALE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CATALASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vainshtein, B.K, Melik-Adamyan, W.R, Barynin, V.V, Vagin, A.A, Grebenko, A.I. | | Deposit date: | 1983-02-24 | | Release date: | 1983-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structure of catalase from Penicillium vitale at 2.0 A resolution.
J.Mol.Biol., 188, 1986
|
|
5O61
 
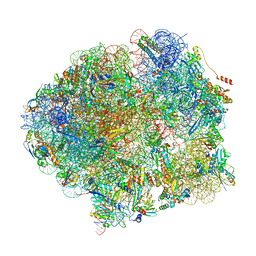 | | The complete structure of the Mycobacterium smegmatis 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hentschel, J, Burnside, C, Mignot, I, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2017-06-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | The Complete Structure of the Mycobacterium smegmatis 70S Ribosome.
Cell Rep, 20, 2017
|
|
5O0S
 
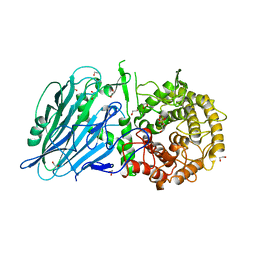 | | Crystal structure of txGH116 (beta-glucosidase from Thermoanaerobacterium xylolyticum) in complex with unreacted beta Cyclophellitol Cyclosulfate probe ME711 | | Descriptor: | (3~{a}~{S},4~{R},5~{S},6~{R},7~{R},7~{a}~{R})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydrobenzo[d][1,3,2]dioxathiole-4,5,6-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Wu, L, Offen, W.A, Breen, I.Z, Davies, G.J. | | Deposit date: | 2017-05-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
1H9H
 
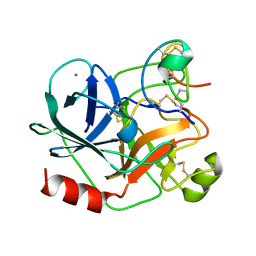 | | COMPLEX OF EETI-II WITH PORCINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Wentzel, A, Kolmar, H, Uson, I. | | Deposit date: | 2001-03-12 | | Release date: | 2004-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3FXQ
 
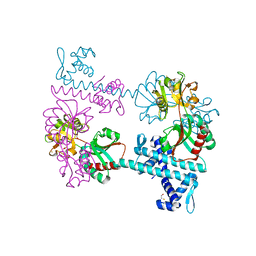 | | Crystal structure of the LysR-type transcriptional regulator TsaR | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Kikhney, A, Svergun, D, Uson, I. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
5NS6
 
 | | Crystal structure of beta-glucosidase BglM-G1 from marine metagenome | | Descriptor: | Beta-glucosidase, GLYCEROL, SULFATE ION | | Authors: | Mhaindarkar, D.C, Gasper, R, Lupilova, N, Leichert, L.I, Hofmann, E. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Loss of a conserved salt bridge in bacterial glycosyl hydrolase BgIM-G1 improves substrate binding in temperate environments.
Commun Biol, 1, 2018
|
|
6ZLB
 
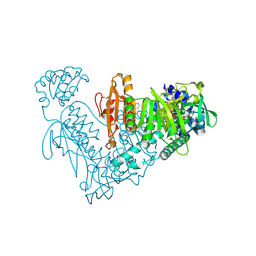 | | Thioredoxin glutathione reductase from Schistosoma mansoni in complex with Indole-3-carbinol | | Descriptor: | 1~{H}-indol-3-ylmethanol, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Silvestri, I, Williams, D.L, Angelucci, F. | | Deposit date: | 2020-06-30 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the Surface of a Parasite Drug Target Thioredoxin Glutathione Reductase Using Small Molecule Fragments.
Acs Infect Dis., 7, 2021
|
|
1SB6
 
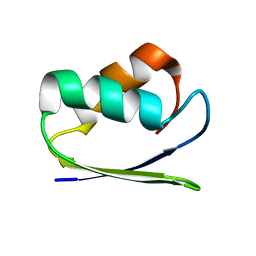 | | Solution structure of a cyanobacterial copper metallochaperone, ScAtx1 | | Descriptor: | copper chaperone ScAtx1 | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.C, Borrelly, G.P, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-02-10 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a Cyanobacterial Metallochaperone: INSIGHT INTO AN ATYPICAL COPPER-BINDING MOTIF.
J.Biol.Chem., 279, 2004
|
|
5NZG
 
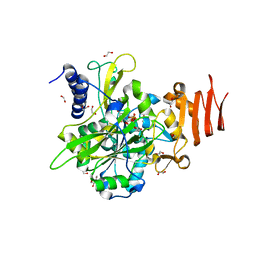 | | Crystal structure of UDP-glucose pyrophosphorylase S374W mutant from Leishmania major in complex with UDP-glucose | | Descriptor: | 1,2-ETHANEDIOL, UDP-glucose pyrophosphorylase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
2D4U
 
 | | Crystal Structure of the ligand binding domain of the bacterial serine chemoreceptor Tsr | | Descriptor: | Methyl-accepting chemotaxis protein I | | Authors: | Imada, K, Tajima, H, Namba, K, Sakuma, M, Homma, M, Kawagishi, I. | | Deposit date: | 2005-10-24 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ligand specificity determined by differentially arranged common ligand-binding residues in the bacterial amino acid chemoreceptors Tsr and Tar.
J.Biol.Chem., 2011
|
|
2FTK
 
 | | berylloflouride Spo0F complex with Spo0B | | Descriptor: | MAGNESIUM ION, Sporulation initiation phosphotransferase B, Sporulation initiation phosphotransferase F | | Authors: | Varughese, K.I. | | Deposit date: | 2006-01-24 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The Crystal Structure of Beryllofluoride Spo0F in Complex with the Phosphotransferase Spo0B Represents a Phosphotransfer Pretransition State.
J.Bacteriol., 188, 2006
|
|
3FXR
 
 | | Crystal structure of TsaR in complex with sulfate | | Descriptor: | ASCORBIC ACID, GLYCEROL, LysR type regulator of tsaMBCD, ... | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Kikhney, A, Svergun, D, Uson, I. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
1HF4
 
 | | STRUCTURAL EFFECTS OF MONOVALENT ANIONS ON POLYMORPHIC LYSOZYME CRYSTALS | | Descriptor: | LYSOZYME, NITRATE ION, SODIUM ION | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 2000-11-29 | | Release date: | 2001-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Effects of Monovalent Anions on Polymorphic Lysozyme Crystals
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6QNP
 
 | |
7QVO
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with guanidine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
5OBW
 
 | | Mycoplasma genitalium DnaK-NBD in complex with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein DnaK, GLYCEROL, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
