2VDC
 
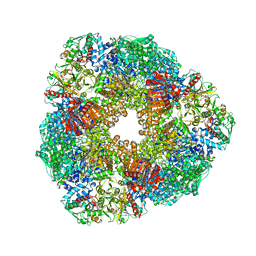 | | THE 9.5 A RESOLUTION STRUCTURE OF GLUTAMATE SYNTHASE FROM CRYO-ELECTRON MICROSCOPY AND ITS OLIGOMERIZATION BEHAVIOR IN SOLUTION: FUNCTIONAL IMPLICATIONS. | | Descriptor: | 2-OXOGLUTARIC ACID, FE3-S4 CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Cottevieille, M, Larquet, E, Jonic, S, Petoukhov, M.V, Caprini, G, Paravisi, S, Svergun, D.I, Vanoni, M.A, Boisset, N. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | The Subnanometer Resolution Structure of the Glutamate Synthase 1.2-Mda Hexamer by Cryoelectron Microscopy and its Oligomerization Behavior in Solution: Functional Implications.
J.Biol.Chem., 283, 2008
|
|
1VZ5
 
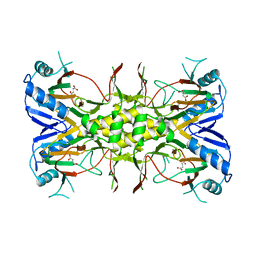 | | Succinate Complex of AtsK | | Descriptor: | PUTATIVE ALKYLSULFATASE ATSK, SUCCINIC ACID | | Authors: | Mueller, I, Stueckl, A.C, Uson, I, Kertesz, M. | | Deposit date: | 2004-05-14 | | Release date: | 2004-11-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Succinate Complex Crystal Structures of the Alpha-Ketoglutarate-Dependent Dioxygenase Atsk: Steric Aspects of Enzyme Self-Hydroxylation
J.Biol.Chem., 280, 2005
|
|
7RTM
 
 | | Cryo-EM Structure of the Sodium-driven Chloride/Bicarbonate Exchanger NDCBE (SLC4A8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Wang, W.G, Tsirulnikov, K, Zhekova, H, Kayik, G, Muhammad-Khan, H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2021-08-13 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the sodium-driven chloride/bicarbonate exchanger NDCBE.
Nat Commun, 12, 2021
|
|
2HD0
 
 | |
6ZT9
 
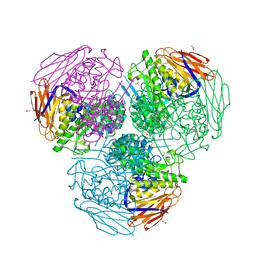 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT8
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | Alpha-L-arabinofuranosidase, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT6
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6ZT7
 
 | | X-ray structure of mutated arabinofuranosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tandrup, T, Lo Leggio, L, Zhao, J, Bissaro, B, Barbe, S, Andre, I, Dumon, C, O'Donohue, M.J, Faure, R. | | Deposit date: | 2020-07-17 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the determinants of the transglycosylation/hydrolysis partition in a retaining alpha-l-arabinofuranosidase.
N Biotechnol, 62, 2021
|
|
6XG4
 
 | | X-ray structure of Escherichia coli dihydrofolate reductase L28R mutant in complex with trimethoprim | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
6XG5
 
 | | X-ray structure of Escherichia coli dihydrofolate reductase in complex with trimethoprim | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Gaszek, I.K, Manna, M.S, Borek, D, Toprak, E. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A trimethoprim derivative impedes antibiotic resistance evolution.
Nat Commun, 12, 2021
|
|
7RDJ
 
 | | Crystal structure of PCDN-16B, an anti-HIV antibody from the PCDN bnAb lineage (non-cysteinylated state) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PCDN-16B Fab heavy chain, ... | | Authors: | Omorodion, O, Wilson, I.A. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Biochemical Characterization of Cysteinylation in Broadly Neutralizing Antibodies to HIV-1.
J.Mol.Biol., 433, 2021
|
|
3FVR
 
 | |
7RDL
 
 | |
7RDM
 
 | |
7JGI
 
 | | NMR structure of the cNTnC-cTnI chimera bound to A7 | | Descriptor: | 7-{[(5-chloronaphthalen-1-yl)sulfonyl]amino}heptanoic acid, CALCIUM ION, Troponin C, ... | | Authors: | Cai, F, Robertson, I.M, Kampourakis, T, Klein, B.A, Sykes, B.D. | | Deposit date: | 2020-07-19 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Role of Electrostatics in the Mechanism of Cardiac Thin Filament Based Sensitizers.
Acs Chem.Biol., 15, 2020
|
|
1JVO
 
 | | Azurin dimer, crosslinked via disulfide bridge | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | van Amsterdam, I.M.C, Ubbink, M, Einsle, O, Messerschmidt, A, Merli, A, Cavazzini, D, Rossi, G.L, Canters, G.W. | | Deposit date: | 2001-08-30 | | Release date: | 2002-01-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dramatic modulation of electron transfer in protein complexes by crosslinking
Nat.Struct.Biol., 9, 2002
|
|
5NDK
 
 | | Crystal structure of aminoglycoside TC007 co-crystallized with 70S ribosome from Thermus thermophilus, three tRNAs and mRNA | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7RDK
 
 | | Crystal structure of PCDN-16B, an anti-HIV antibody from the PCDN bnAb lineage (cysteinylated state) | | Descriptor: | 1,2-ETHANEDIOL, CYSTEINE, GLYCEROL, ... | | Authors: | Omorodion, O, Wilson, I.A. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and Biochemical Characterization of Cysteinylation in Broadly Neutralizing Antibodies to HIV-1.
J.Mol.Biol., 433, 2021
|
|
7RTQ
 
 | | Sterol 14alpha demethylase (CYP51) from Naegleria fowleri in complex with an inhibitor R)-N-(1-(3,4'-difluorobiphenyl-4-yl)-2-(1H-imidazol-1-yl)ethyl)-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide | | Descriptor: | N-[(1R)-2-(1H-imidazol-1-yl)-1-(3,4',5-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-4-(5-phenyl-1,3,4-oxadiazol-2-yl)benzamide, PROTOPORPHYRIN IX CONTAINING FE, Protein CYP51 | | Authors: | Lepesheva, G.I, Hargrove, T.Y, Wawrzak, Z. | | Deposit date: | 2021-08-13 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Relaxed Substrate Requirements of Sterol 14 alpha-Demethylase from Naegleria fowleri Are Accompanied by Resistance to Inhibition.
J.Med.Chem., 64, 2021
|
|
6ZPL
 
 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor, sybody Sb_GlyT1#7 and bound Na and Cl ions. | | Descriptor: | CHLORIDE ION, Endoglucanase H, SODIUM ION, ... | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.945 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
7RB1
 
 | | Isocitrate Lyase-1 from Mycobacterium tuberculosis covalently modified by 5-descarboxy-5-nitro-D-isocitric acid | | Descriptor: | (3E)-3-(hydroxyimino)propanoic acid, GLYCEROL, GLYOXYLIC ACID, ... | | Authors: | Krieger, I.V, Mellott, D, Meek, T, Sacchettini, J.C. | | Deposit date: | 2021-07-05 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism-Based Inactivation of Mycobacterium tuberculosis Isocitrate Lyase 1 by (2 R ,3 S )-2-Hydroxy-3-(nitromethyl)succinic acid.
J.Am.Chem.Soc., 143, 2021
|
|
8K3F
 
 | |
6ZS1
 
 | | Chaetomium thermophilum CuZn-superoxide dismutase | | Descriptor: | COPPER (II) ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Papageorgiou, A.C, Mohsin, I. | | Deposit date: | 2020-07-15 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of a Cu,Zn Superoxide Dismutase From the Thermophilic Fungus Chaetomium thermophilum.
Protein Pept.Lett., 28, 2021
|
|
1RXU
 
 | | E. coli uridine phosphorylase: thymidine phosphate complex | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, THYMIDINE, ... | | Authors: | Caradoc-Davies, T.T, Cutfield, S.M, Lamont, I.L, Cutfield, J.F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium
J.Mol.Biol., 337, 2004
|
|
1S3Q
 
 | | Crystal structures of a novel open pore ferritin from the hyperthermophilic Archaeon Archaeoglobus fulgidus | | Descriptor: | ZINC ION, ferritin | | Authors: | Johnson, E, Cascio, D, Sawaya, M, Schroeder, I. | | Deposit date: | 2004-01-13 | | Release date: | 2005-04-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of a tetrahedral open pore ferritin from the hyperthermophilic archaeon Archaeoglobus fulgidus.
Structure, 13, 2005
|
|
