5W6A
 
 | | HLA-C*06:02 presenting ARTELYRSL | | Descriptor: | ALA-ARG-THR-GLU-LEU-TYR-ARG-SER-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mobbs, J.I, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule.
J. Biol. Chem., 292, 2017
|
|
6NQL
 
 | | Crystal structure of fast switching M159T mutant of fluorescent protein Dronpa (Dronpa2), Y63(3-ClY) | | Descriptor: | Fluorescent protein Dronpa, TRIETHYLENE GLYCOL | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2019-01-21 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Electrostatic control of photoisomerization pathways in proteins.
Science, 367, 2020
|
|
1XE8
 
 | | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly roll fold. | | Descriptor: | ADENINE, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhou, C.-Z, Meyer, P, Quevillon-Cheruel, S, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Leulliot, N, Sorel, I, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2004-09-09 | | Release date: | 2005-01-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly-roll fold
Protein Sci., 14, 2005
|
|
3N02
 
 | | Thaumatic crystals grown in loops/micromounts | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Mathes, I.I. | | Deposit date: | 2010-05-13 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diffraction study of protein crystals grown in cryoloops and micromounts.
J.Appl.Crystallogr., 43, 2010
|
|
3MXB
 
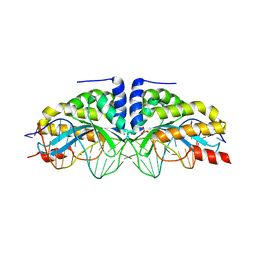 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*CP*TP*GP*GP*CP*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*GP*AP*GP*AP*AP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*TP*CP*TP*CP*AP*GP*GP*TP*AP*CP*CP*TP*CP*AP*GP*CP*CP*AP*GP*A)-3'), ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Montoya, G. | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus.
Nucleic Acids Res., 39, 2011
|
|
2CAK
 
 | | 1.27Angstrom Structure of Rusticyanin from Thiobacillus ferrooxidans | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Barrett, M.L, Harvey, I, Sundararajan, M, Surendran, R, Hall, J.F, Ellis, M.J, Hough, M.A, Strange, R.W, Hillier, I.H, Hasnain, S.S. | | Deposit date: | 2005-12-21 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Atomic Resolution Crystal Structures, Exafs, and Quantum Chemical Studies of Rusticyanin and its Two Mutants Provide Insight Into its Unusual Properties.
Biochemistry, 45, 2006
|
|
2Q9F
 
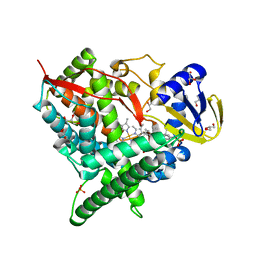 | | Crystal structure of human cytochrome P450 46A1 in complex with cholesterol-3-sulphate | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, Cytochrome P450 46A1, GLYCEROL, ... | | Authors: | White, M.A, Mast, N.V, Johnson, E.F, Stout, C.D, Pikuleva, I.A. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3RJ1
 
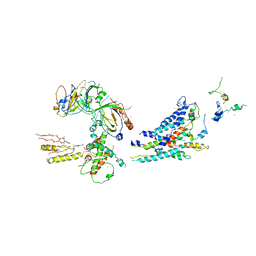 | | Architecture of the Mediator Head module | | Descriptor: | Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, Mediator of RNA polymerase II transcription subunit 18, ... | | Authors: | Imasaki, T, Calero, G, Cai, G, Tsai, K.L, Yamada, K, Cardelli, F, Erdjument-Bromage, H, Tempst, P, Berger, I, Kornberg, G.L, Asturias, F.J, Kornberg, R.D, Takagi, Y. | | Deposit date: | 2011-04-14 | | Release date: | 2011-07-27 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Architecture of the Mediator head module.
Nature, 475, 2011
|
|
2PPE
 
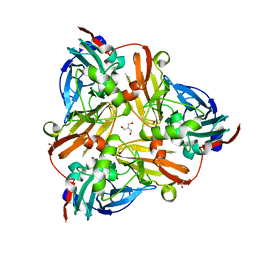 | | Reduced H145A mutant of AfNiR exposed to NO | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, COPPER (I) ION, ... | | Authors: | Tocheva, E.I, Murphy, M.E.P. | | Deposit date: | 2007-04-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stable copper-nitrosyl formation by nitrite reductase in either oxidation state
Biochemistry, 46, 2007
|
|
1OIH
 
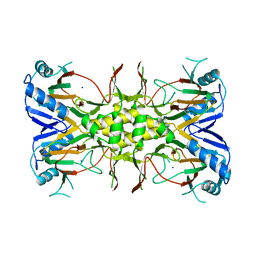 | | Crystal structure of the alkylsulfatase AtsK, a non-heme Fe(II) alphaketoglutarate dependent Dioxygenase | | Descriptor: | PUTATIVE ALKYLSULFATASE ATSK, SODIUM ION | | Authors: | Mueller, I, Kahnert, A, Pape, T, Dierks, T, Meyer-Klauke, W, Kertesz, M, Uson, I. | | Deposit date: | 2003-06-18 | | Release date: | 2004-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Alkylsulfatase Atsk: Insights Into the Catalytic Mechanism of the Fe(II) -Ketoglutarate-Dependent Dioxygenase Superfamily
Biochemistry, 42, 2004
|
|
4H3D
 
 | | 1.95 Angstrom Crystal Structure of of Type I 3-Dehydroquinate Dehydratase (aroD) from Clostridium difficile with Covalent Modified Comenic Acid. | | Descriptor: | 3-dehydroquinate dehydratase, 5-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid, ACETATE ION, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Duban, M.-E, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of of Type I 3-Dehydroquinate Dehydratase (aroD) from Clostridium difficile with Covalent Modified Comenic Acid.
TO BE PUBLISHED
|
|
2ER0
 
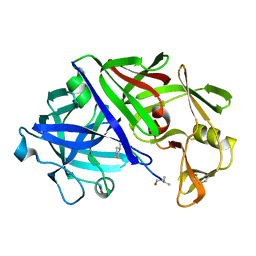 | | X-RAY STUDIES OF ASPARTIC PROTEINASE-STATINE INHIBITOR COMPLEXES | | Descriptor: | ENDOTHIAPEPSIN, L364,099 | | Authors: | Cooper, J.B, Foundling, S.I, Boger, J, Blundell, T.L. | | Deposit date: | 1990-10-20 | | Release date: | 1991-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray studies of aspartic proteinase-statine inhibitor complexes.
Biochemistry, 28, 1989
|
|
4LG2
 
 | | Crystal structure of Reston Ebola virus VP35 RNA binding domain bound to 12-bp dsRNA | | Descriptor: | Polymerase cofactor, dsRNA | | Authors: | Bale, S, Julien, J.-P, Bornholdt, Z.A, Krois, A.S, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2013-06-27 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ebolavirus VP35 coats the backbone of double-stranded RNA for interferon antagonism.
J.Virol., 87, 2013
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
2RML
 
 | | Solution structure of the N-terminal soluble domains of Bacillus subtilis CopA | | Descriptor: | Copper-transporting P-type ATPase copA | | Authors: | Singleton, C, Banci, L, Bertini, I, Ciofi-Baffoni, S, Tenori, L, Kihlken, M.A, Boetzel, R, Le Brun, N.E. | | Deposit date: | 2007-10-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Cu(I)-binding properties of the N-terminal soluble domains of Bacillus subtilis CopA
Biochem.J., 411, 2008
|
|
1JWW
 
 | | NMR characterization of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis | | Descriptor: | Potential copper-transporting ATPase | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, D'Onofrio, M, Gonnelli, L, Marhuenda-Egea, F, Ruiz-Duenas, F.J. | | Deposit date: | 2001-09-05 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a potential copper-translocating P-type ATPase from Bacillus subtilis in the apo and Cu(I) loaded states.
J.Mol.Biol., 317, 2002
|
|
1FPC
 
 | | ACTIVE SITE MIMETIC INHIBITION OF THROMBIN | | Descriptor: | Hirudin, amino{[(4S)-4-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)-5-(4-ethylpiperidin-1-yl)-5-oxopentyl]amino}methaniminium, thrombin | | Authors: | Tulinsky, A, Mathews, I.I. | | Deposit date: | 1994-10-16 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-site mimetic inhibition of thrombin.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1XZ0
 
 | | Crystal structure of CD1a in complex with a synthetic mycobactin lipopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(HYDROXY-HEXADECANOYL-AMINO)-2-{[(4S)-2-(2-HYDROXY-PHENYL)-4,5-DIHYDRO-OXAZOLE-4-CARBONYL]-AMINO}-HEXANOIC ACID 2-[(3S)-1-(TERT-BUTYL-DIPHENYL-SILANYLOXY)-2-OXO-AZEPAN-3-YLCARBAMOYL]-(1S)-1-METHYL-ETHYL ESTER, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Crispin, M.D, Bowden, T.A, Young, D.C, Cheng, T.Y, Hu, J, Costello, C.E, Miller, M.J, Moody, D.B, Wilson, I.A. | | Deposit date: | 2004-11-11 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism of Lipopeptide Presentation by CD1a.
Immunity, 22, 2005
|
|
4WX1
 
 | |
2Y9N
 
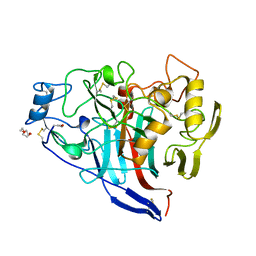 | | Cellobiohydrolase I Cel7A from Trichoderma harzianum at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EXOGLUCANASE 1, TRIETHYLENE GLYCOL | | Authors: | Textor, L.C, Colussi, F, Serpa, V, Squina, F, Pereira Jr, N, Polikarpov, I. | | Deposit date: | 2011-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Cellobiohydrolase I from Trichoderma Harzianum: Structural and Enzymatic Characterization
To be Published
|
|
8EVN
 
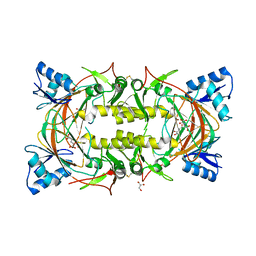 | |
5OXX
 
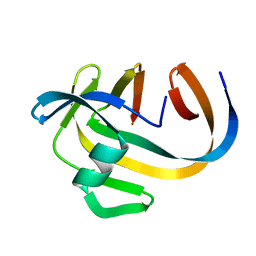 | | Crystal structure of NeqN/NeqC complex from Nanoarcheaum equitans at 1.7A | | Descriptor: | NEQ068, NEQ528 | | Authors: | Aparicio, D, Perez-Luque, R, Ribo, M, Fita, I. | | Deposit date: | 2017-09-07 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Insights into Subunits Assembly and the Oxyester Splicing Mechanism of Neq pol Split Intein.
Cell Chem Biol, 25, 2018
|
|
5OYA
 
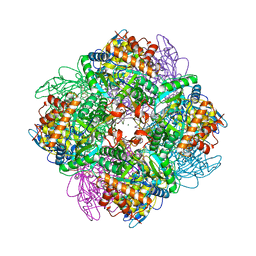 | | Unusual posttranslational modifications revealed in crystal structures of diatom Rubisco. | | Descriptor: | 1,2-ETHANEDIOL, 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Valegard, K, Andersson, I. | | Deposit date: | 2017-09-08 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analyses of Rubisco from arctic diatom species reveal unusual posttranslational modifications.
J. Biol. Chem., 293, 2018
|
|
4X35
 
 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | Deposit date: | 2014-11-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
6QKD
 
 | | CRYSTAL STRUCTURE OF vhh-based FAB-fragment of antibody BCD-085 | | Descriptor: | CHLORIDE ION, FAB HEAVY CHAIN, FAB LIGHT CHAIN, ... | | Authors: | Kostareva, O.S, Kolyadenko, I.A, Ulitin, A.B, Ekimova, V.M, Evdokimov, S.R, Garber, M.B, Tishchenko, T.V, Gabdulkhakov, A.G. | | Deposit date: | 2019-01-29 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fab Fragment of VHH-Based Antibody Netakimab: Crystal Structure and Modeling Interaction with Cytokine IL-17A
Crystals, 2019
|
|
