6CSZ
 
 | | TFE-induced NMR structure of a novel bioactive peptide (PaDBS1R3) derived from a Pyrobaculum aerophilum ribosomal protein (L39e) | | 分子名称: | PaDBS1R3 | | 著者 | Cardoso, M.H, Chan, L.Y, Candido, E.S, Torres, M.T, Oshiro, K.G.N, Silva, I.C, Goncalves, S, Buccini, D.F, Lu, T, Santos, N.C, de la Fuente-Nunez, C, Craik, D.J, Franco, O.L. | | 登録日 | 2018-03-21 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An N-capping asparagine-lysine-proline (NKP) motif contributes to a hybrid flexible/stable multifunctional peptide scaffold
Chem Sci, 13, 2022
|
|
3QRO
 
 | | HIV-1 protease (mutant Q7K L33I L63I) in complex with a three-armed pyrrolidine-based inhibitor | | 分子名称: | 4-({(3S,4S)-4-[(3,5-dihydroxybenzyl)amino]pyrrolidin-3-yl}[4-(trifluoromethyl)benzyl]sulfamoyl)benzamide, CHLORIDE ION, DITHIANE DIOL, ... | | 著者 | Lindemann, I, Heine, A, Klebe, G. | | 登録日 | 2011-02-18 | | 公開日 | 2012-02-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.616 Å) | | 主引用文献 | Design of a series of novel three-armed pyrrolidine-based inhibitors for HIV-1 protease
To be Published
|
|
6CSK
 
 | | TFE-induced NMR structure of a novel bioactive peptide (PaDBS1R2) derived from a Pyrobaculum aerophilum ribosomal protein (L39e) | | 分子名称: | PaDBS1R2 | | 著者 | Cardoso, M.H, Chan, L.Y, Candido, E.S, Torres, M.T, Oshiro, K.G.N, Silva, I.C, Goncalves, S, Buccini, D.F, Lu, T, Santos, N.C, de la Fuente-Nunez, C, Craik, D.J, Franco, O.L. | | 登録日 | 2018-03-20 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An N-capping asparagine-lysine-proline (NKP) motif contributes to a hybrid flexible/stable multifunctional peptide scaffold
Chem Sci, 13, 2022
|
|
2OP6
 
 | | Peptide-binding domain of Heat shock 70 kDa protein D precursor from C.elegans | | 分子名称: | Heat shock 70 kDa protein D | | 著者 | Osipiuk, J, Duggan, E, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-01-26 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | X-ray structure of peptide-binding domain of Heat shock 70 kDa protein D precursor from C.elegans
To be Published
|
|
1TQG
 
 | | CheA phosphotransferase domain from Thermotoga maritima | | 分子名称: | Chemotaxis protein cheA | | 著者 | Quezada, C.M, Gradinaru, C, Simon, M.I, Bilwes, A.M, Crane, B.R. | | 登録日 | 2004-06-17 | | 公開日 | 2004-09-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | Helical Shifts Generate Two Distinct Conformers in the Atomic Resolution Structure of the CheA Phosphotransferase Domain from Thermotoga maritima.
J.Mol.Biol., 341, 2004
|
|
5HR1
 
 | | Crystal structure of thioredoxin L107A mutant | | 分子名称: | COPPER (II) ION, Thioredoxin-1 | | 著者 | Noguera, M.E, Vazquez, D.S, Howard, E.I, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | 登録日 | 2016-01-22 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.144 Å) | | 主引用文献 | Structural variability of E. coli thioredoxin captured in the crystal structures of single-point mutants.
Sci Rep, 7, 2017
|
|
4AFM
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens. | | 分子名称: | ACETATE ION, ENDOGLUCANASE CEL5A, GLYCEROL | | 著者 | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | 登録日 | 2012-01-19 | | 公開日 | 2012-12-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan.
J.Biol.Chem., 288, 2013
|
|
2OP4
 
 | | Crystal Structure of Quorum-Quenching Antibody 1G9 | | 分子名称: | 1,2-ETHANEDIOL, Murine Antibody Fab RS2-1G9 IGG1 Heavy Chain, Murine Antibody Fab RS2-1G9 Lambda Light Chain | | 著者 | Kirchdoerfer, R.N, Debler, E.W, Wilson, I.A. | | 登録日 | 2007-01-26 | | 公開日 | 2007-05-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal Structures of a Quorum-quenching Antibody.
J.Mol.Biol., 368, 2007
|
|
4A68
 
 | | Mutations in the neighbourhood of CotA-laccase trinuclear site: D116N mutant | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, ... | | 著者 | Silva, C.S, Lindley, P.F, Bento, I. | | 登録日 | 2011-10-31 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Role of Asp116 in the Reductive Cleavage of Dioxygen to Water in Cota Laccase: Assistance During the Proton Transfer Mechanism
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2Y4V
 
 | |
6D7V
 
 | | Crystal Structure of Rat TRPV6*- in complex with brominated 2-Aminoethoxydiphenyl borate (2-APB-Br) | | 分子名称: | 2-aminoethyl (4-bromophenyl)phenylborinate, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | 著者 | Singh, A.K, Saotome, K, McGoldrick, L.L, Sobolevsky, A.I. | | 登録日 | 2018-04-25 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB.
Nat Commun, 9, 2018
|
|
2Y33
 
 | |
4QQV
 
 | | Extracellular domains of mouse IL-3 beta receptor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-3 receptor class 2 subunit beta | | 著者 | Jackson, C.J, Young, I.G, Murphy, J.M, Carr, P.D, Ewens, C.L, Dai, J, Ollis, D.L. | | 登録日 | 2014-06-30 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.45 Å) | | 主引用文献 | Crystal structure of the mouse interleukin-3 beta-receptor: insights into interleukin-3 binding and receptor activation.
Biochem.J., 463, 2014
|
|
4ADL
 
 | | Crystal structures of Rv1098c in complex with malate | | 分子名称: | (2S)-2-hydroxybutanedioic acid, FUMARATE HYDRATASE CLASS II | | 著者 | Mechaly, A.E, Haouz, A, Miras, I, Weber, P, Shepard, W, Cole, S, Alzari, P.M, Bellinzoni, M. | | 登録日 | 2011-12-26 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
2OQK
 
 | | Crystal structure of putative Cryptosporidium parvum translation initiation factor eIF-1A | | 分子名称: | Putative translation initiation factor eIF-1A, SULFATE ION | | 著者 | Dong, A, Lew, J, Zhao, Y, Hassanali, A, Lin, L, Qiu, W, Brokx, S.J, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Altamentova, S, Structural Genomics Consortium (SGC) | | 登録日 | 2007-01-31 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of putative Cryptosporidium parvum translation initiation factor eIF-1A
To be Published
|
|
4A6V
 
 | | X-ray structures of oxazole hydroxamate EcMetAp-Mn complexes | | 分子名称: | CARBONATE ION, MANGANESE (II) ION, METHIONINE AMINOPEPTIDASE, ... | | 著者 | Huguet, F, Melet, A, AlvesdeSousa, R, Lieutaud, A, Chevalier, J, Deschamps, P, Tomas, A, Leulliot, N, Pages, J.M, Artaud, I. | | 登録日 | 2011-11-09 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Hydroxamic Acids as Potent Inhibitors of Fe(II) and Mn(II) E. Coli Methionine Aminopeptidase: Biological Activities and X-Ray Structures of Oxazole Hydroxamate-Ecmetap-Mn Complexes.
Chemmedchem, 7, 2012
|
|
4A9U
 
 | | CRYSTAL STRUCTURE OF HUMAN CHK2 IN COMPLEX WITH BENZIMIDAZOLE CARBOXAMIDE INHIBITOR | | 分子名称: | 1,2-ETHANEDIOL, 2-{4-[(1-BENZYLPIPERIDIN-4-YL)METHOXY]PHENYL}-1H-BENZIMIDAZOLE-6-CARBOXAMIDE, CHLORIDE ION, ... | | 著者 | Matijssen, C, Silva-Santisteban, M.C, Westwood, I.M, Siddique, S, Choi, V, Sheldrake, P, van Montfort, R.L.M, Blagg, J. | | 登録日 | 2011-11-28 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Benzimidazole Inhibitors of the Protein Kinase Chk2: Clarification of the Binding Mode by Flexible Side Chain Docking and Protein-Ligand Crystallography
Bioorg.Med.Chem., 20, 2012
|
|
6D7S
 
 | |
4AHT
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | 分子名称: | 1,2-ETHANEDIOL, 1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | 著者 | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | 登録日 | 2012-02-07 | | 公開日 | 2012-12-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
4AJZ
 
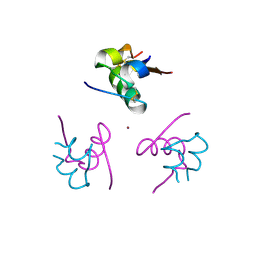 | | Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin | | 分子名称: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | 著者 | Steensgaard, D.B, Schluckebier, G, Strauss, H.M, Norrman, M, Thomsen, J.K, Friderichsen, A.V, Havelund, S, Jonassen, I. | | 登録日 | 2012-02-21 | | 公開日 | 2013-01-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Ligand Controlled Assembly of Hexamers, Dihexamers, and Linear Multihexamer Structures by the Engineered Acylated Insulin Degludec.
Biochemistry, 52, 2013
|
|
3GNU
 
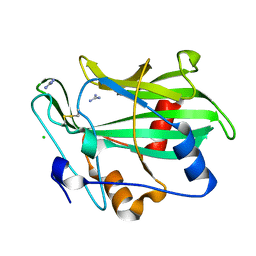 | | Toxin fold as basis for microbial attack and plant defense | | 分子名称: | 25 kDa protein elicitor, CHLORIDE ION, GUANIDINE | | 著者 | Ottmann, C, Luberacki, B, Kuefner, I, Koch, W, Brunner, F, Weyand, M, Mattinen, L, Pirhonen, M, Anderluh, G, Seitz, H.U, Nuernberger, T, Oecking, C. | | 登録日 | 2009-03-18 | | 公開日 | 2009-06-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A common toxin fold mediates microbial attack and plant defense
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1TZW
 
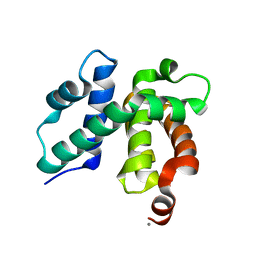 | | T. maritima NusB, P3121, Form 2 | | 分子名称: | CALCIUM ION, N utilization substance protein B homolog | | 著者 | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | 登録日 | 2004-07-12 | | 公開日 | 2004-08-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
2OUG
 
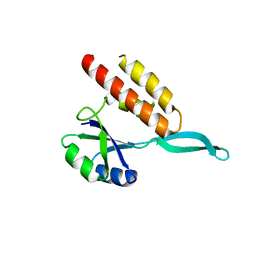 | |
6D8X
 
 | | PPAR gamma LBD complexed with the agonist GW1929 | | 分子名称: | (2~{S})-3-[4-[2-[methyl(pyridin-2-yl)amino]ethoxy]phenyl]-2-[[2-(phenylcarbonyl)phenyl]amino]propanoic acid, CITRATE ANION, GLYCEROL, ... | | 著者 | Mou, T.C, Chrisman, I.M, Hughes, T.S, Sprang, S.R. | | 登録日 | 2018-04-27 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | PPAR gamma LBD complexed with the agonist GW1929
To Be Published
|
|
6DB1
 
 | | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica | | 分子名称: | CHLORIDE ION, Putative methyl-accepting chemotaxis protein | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-05-02 | | 公開日 | 2018-05-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica.
To Be Published
|
|
