3WIL
 
 | | Crystal structure of the CK2alpha/compound3 complex | | Descriptor: | Casein kinase II subunit alpha, {[(2Z)-2-(3,4-dimethoxybenzylidene)-3-oxo-2,3-dihydro-1-benzofuran-6-yl]oxy}acetic acid | | Authors: | Kinoshita, T, Nakanishi, I. | | Deposit date: | 2013-09-18 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of protein kinase CK2 inhibitors using solvent dipole ordering virtual screening
To be Published
|
|
4IH8
 
 | | Crystal structure of TgCDPK1 with inhibitor bound | | Descriptor: | 4-Amino-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-7-yl-cyclopentane, Calmodulin-domain protein kinase 1 | | Authors: | El Bakkouri, M, Tempel, W, Crandall, I, Massad, T, Loppnau, P, Graslund, S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kain, K, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.877 Å) | | Cite: | Crystal structure of TgCDPK1 with inhibitor bound
To be Published
|
|
4IFK
 
 | | Arginines 51 and 239* from a Neighboring Subunit are Essential for Catalysis in a Zinc-dependent Decarboxylase | | Descriptor: | 2-amino-3-carboxymuconate 6-semialdehyde decarboxylase, MAGNESIUM ION, ZINC ION | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | The power of two: arginine 51 and arginine 239* from a neighboring subunit are essential for catalysis in alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase.
J.Biol.Chem., 288, 2013
|
|
4A9T
 
 | | CRYSTAL STRUCTURE OF HUMAN CHK2 IN COMPLEX WITH BENZIMIDAZOLE CARBOXAMIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-{[1-(4-CHLOROBENZYL)PIPERIDIN-4-YL]METHOXY}PHENYL)-1H-BENZIMIDAZOLE-5-CARBOXAMIDE, NITRATE ION, ... | | Authors: | Matijssen, C, Silva-Santisteban, M.C, Westwood, I.M, Siddique, S, Choi, V, Sheldrake, P, van Montfort, R.L.M, Blagg, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Benzimidazole Inhibitors of the Protein Kinase Chk2: Clarification of the Binding Mode by Flexible Side Chain Docking and Protein-Ligand Crystallography.
Bioorg.Med.Chem., 20, 2012
|
|
3Q7B
 
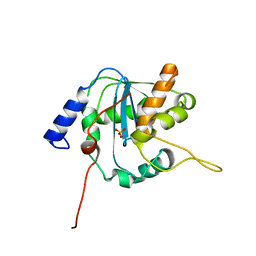 | | Exonuclease domain of Lassa virus nucleoprotein | | Descriptor: | Nucleoprotein, PHOSPHATE ION, ZINC ION | | Authors: | Hastie, K.M, Kimberlin, C.R, Zandonatti, M.A, MacRae, I.J, Saphire, E.O. | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3' to 5' exonuclease activity essential for immune suppression.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4A67
 
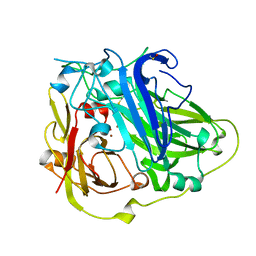 | | Mutations in the neighbourhood of CotA-laccase trinuclear site: D116E mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, SPORE COAT PROTEIN A | | Authors: | Silva, C.S, Lindley, P.F, Bento, I. | | Deposit date: | 2011-10-31 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Role of Asp116 in the Reductive Cleavage of Dioxygen to Water in Cota Laccase: Assistance During the Proton Transfer Mechanism
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4JHN
 
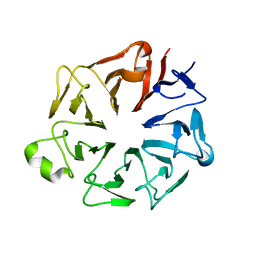 | | The crystal structure of the RPGR RCC1-like domain | | Descriptor: | X-linked retinitis pigmentosa GTPase regulator | | Authors: | Waetzlich, D, Vetter, I, Wittinghofer, A, Ismail, S. | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The interplay between RPGR, PDE-delta and Arl2/3 regulate the ciliary targeting of farnesylated cargo.
Embo Rep., 14, 2013
|
|
4JM7
 
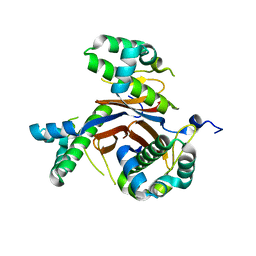 | | 1.82 Angstrom resolution crystal structure of holo-(acyl-carrier-protein) synthase (acpS) from Staphylococcus aureus | | Descriptor: | Holo-[acyl-carrier-protein] synthase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Structural characterization and comparison of three acyl-carrier-protein synthases from pathogenic bacteria.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4JNH
 
 | |
3QK6
 
 | | Crystal structure of Escherichia coli PhnD | | Descriptor: | PhnD, subunit of alkylphosphonate ABC transporter, UNKNOWN LIGAND | | Authors: | Alicea, I, Schreiter, E.R. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-12 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Escherichia coli Phosphonate Binding Protein PhnD and Rationally Optimized Phosphonate Biosensors.
J.Mol.Biol., 414, 2011
|
|
3WEK
 
 | | Crystal structure of the human squalene synthase F288L mutant in complex with presqualene pyrophosphate | | Descriptor: | MAGNESIUM ION, Squalene synthase, {(1R,2R,3R)-2-[(3E)-4,8-dimethylnona-3,7-dien-1-yl]-2-methyl-3-[(1E,5E)-2,6,10-trimethylundeca-1,5,9-trien-1-yl]cyclopropyl}methyl trihydrogen diphosphate | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-07-07 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the catalytic mechanism of human squalene synthase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5COL
 
 | |
3R4Z
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) in complex with alpha-d-galactopyranose from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal, alpha-D-galactopyranose | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
3R6K
 
 | | Crystal Structure of the Capsid P Domain from Norwalk Virus Strain Hiroshima/1999 in complex with HBGA type B (triglycan) | | Descriptor: | 1,2-ETHANEDIOL, VP1 protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Hansman, G.S, Biertumpfel, C, McLellan, J.S, Georgiev, I, Chen, L, Zhou, T, Katayama, K, Kwong, P.D. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
4A6W
 
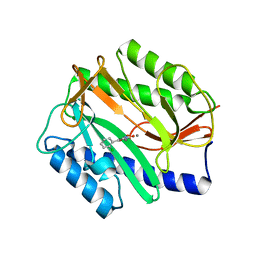 | | X-ray structures of oxazole hydroxamate EcMetAp-Mn complexes | | Descriptor: | 5-(2-chlorophenyl)-N-hydroxy-1,3-oxazole-2-carboxamide, MANGANESE (II) ION, METHIONINE AMINOPEPTIDASE | | Authors: | Huguet, F, Melet, A, AlvesdeSousa, R, Lieutaud, A, Chevalier, J, Deschamps, P, Tomas, A, Leulliot, N, Pages, J.M, Artaud, I. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Hydroxamic Acids as Potent Inhibitors of Fe(II) and Mn(II) E. Coli Methionine Aminopeptidase: Biological Activities and X-Ray Structures of Oxazole Hydroxamate-Ecmetap-Mn Complexes.
Chemmedchem, 7, 2012
|
|
4J2T
 
 | | Inhibitor-bound Ca2+ ATPase | | Descriptor: | (3S,3aR,4S,6S,6aR,7S,8S,9bS)-6-(acetyloxy)-3a,4-bis(butanoyloxy)-3-hydroxy-3,6,9-trimethyl-8-{[(2E)-2-methylbut-2-enoyl]oxy}-2-oxo-2,3,3a,4,5,6,6a,7,8,9b-decahydroazuleno[4,5-b]furan-7-yl octanoate, PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, ... | | Authors: | Paulsen, E.S, Villadsen, J, Tenori, E, Liu, H, Lie, M.A, Bonde, D.F, Bublitz, M, Olesen, C, Autzen, H.E, Dach, I, Sehgal, P, Moller, J.V, Schiott, B, Nissen, P, Christensen, S.B. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Water-mediated interactions influence the binding of thapsigargin to sarco/endoplasmic reticulum calcium adenosinetriphosphatase.
J.Med.Chem., 56, 2013
|
|
4J18
 
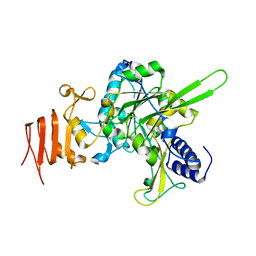 | | Crystal structure of H191L mutant of UDP-glucose pyrophosphorylase from Leishmania major | | Descriptor: | UDP-glucose pyrophosphorylase | | Authors: | Fuehring, J.I, Routier, F.H, Lamerz, A.-C, Baruch, P, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2013-02-01 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Catalytic Mechanism and Allosteric Regulation of UDP-Glucose Pyrophosphorylase from Leishmania major
ACS CATALYSIS, 3, 2013
|
|
4A9R
 
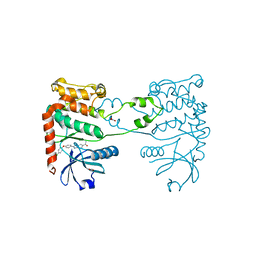 | | CRYSTAL STRUCTURE OF HUMAN CHK2 IN COMPLEX WITH BENZIMIDAZOLE CARBOXAMIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(4-CHLOROPHENOXY)PHENYL]-1H-BENZIMIDAZOLE-6-CARBOXAMIDE, NITRATE ION, ... | | Authors: | Matijssen, C, Silva-Santisteban, M.C, Westwood, I.M, Siddique, S, Choi, V, Sheldrake, P, van Montfort, R.L.M, Blagg, J. | | Deposit date: | 2011-11-28 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Benzimidazole Inhibitors of the Protein Kinase Chk2: Clarification of the Binding Mode by Flexible Side Chain Docking and Protein-Ligand Crystallography
Bioorg.Med.Chem., 20, 2012
|
|
4AIM
 
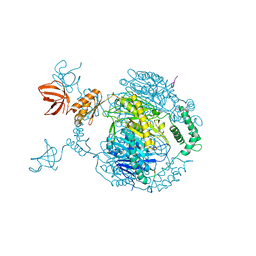 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-02-10 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
4AEM
 
 | | Structural and biochemical characterization of a novel Carbohydrate Binding Module of endoglucanase Cel5A from Eubacterium cellulosolvens | | Descriptor: | ENDOGLUCANASE CEL5A | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan
J.Biol.Chem., 288, 2013
|
|
3U71
 
 | | Crystal Structure Analysis of South African wild type HIV-1 Subtype C Protease | | Descriptor: | HIV-1 Protease | | Authors: | Naicker, P, Fanucchi, S, Achilonu, I.A, Fernandes, M.A, Dirr, H.W, Sayed, Y. | | Deposit date: | 2011-10-13 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure analysis of South African wild type HIV-1 subtype C apo protease
To be Published
|
|
3U7R
 
 | | FerB - flavoenzyme NAD(P)H:(acceptor) oxidoreductase (FerB) from Paracoccus denitrificans | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NADPH-dependent FMN reductase, NONAETHYLENE GLYCOL | | Authors: | Marek, J, Klumpler, T, Sedlacek, V, Kucera, I. | | Deposit date: | 2011-10-14 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Structural and Functional Basis of Catalysis Mediated by NAD(P)H:acceptor Oxidoreductase (FerB) of Paracoccus denitrificans.
Plos One, 9, 2014
|
|
3WGN
 
 | | STAPHYLOCOCCUS AUREUS FTSZ bound with GTP-gamma-S | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Cell division protein FtsZ | | Authors: | Matsui, T, Mogi, N, Tanaka, I, Yao, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop
J.Biol.Chem., 289, 2014
|
|
3WH0
 
 | | Structure of Pin1 Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Lee, C.C, Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3WGK
 
 | | STAPHYLOCOCCUS AUREUS FTSZ T7 mutant substituted for GAG, DeltaT7GAG-GDP | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Han, X, Matsui, T, Yu, J, Tanaka, I, Yao, M. | | Deposit date: | 2013-08-06 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop
J.Biol.Chem., 289, 2014
|
|
