5LIZ
 
 | | The structure of Nt.BspD6I nicking endonuclease with all cysteines mutated by serine residues at 0.19 nm resolution . | | Descriptor: | Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Artyukh, R.I, Perevyazova, T.A, Yunusova, A.K, Popov, A.N, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural features of Cysteine residues mutation of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
6HQ7
 
 | | Structure of EAL Enzyme Bd1971 - cGMP bound form | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, EAL Enzyme Bd1971, MAGNESIUM ION | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2018-09-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase.
Embo J., 38, 2019
|
|
4L92
 
 | | Structure of the RBP from lactococcal phage 1358 in complex with 2 GlcNAc molecules | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor Binding Protein, ... | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
1QE0
 
 | | CRYSTAL STRUCTURE OF APO S. AUREUS HISTIDYL-TRNA SYNTHETASE | | Descriptor: | Histidine--tRNA ligase | | Authors: | Qiu, X, Janson, C.A, Blackburn, M.N, Chohan, I.K, Hibbs, M, Abdel-Meguid, S.S. | | Deposit date: | 1999-07-12 | | Release date: | 2000-07-12 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative structural dynamics and a novel fidelity mechanism in histidyl-tRNA synthetases.
Biochemistry, 38, 1999
|
|
1XYF
 
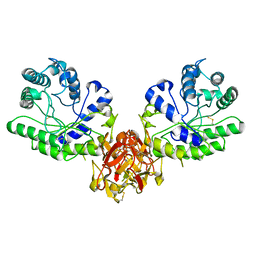 | | ENDO-1,4-BETA-XYLANASE FROM STREPTOMYCES OLIVACEOVIRIDIS | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Fujimoto, Z, Mizuno, H, Kuno, A, Kusakabe, I. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Streptomyces olivaceoviridis E-86 beta-xylanase containing xylan-binding domain.
J.Mol.Biol., 300, 2000
|
|
1QGB
 
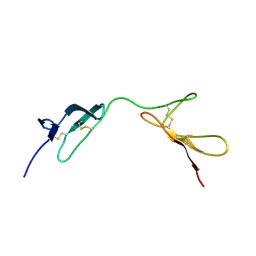 | | SOLUTION STRUCTURE OF THE N-TERMINAL F1 MODULE PAIR FROM HUMAN FIBRONECTIN | | Descriptor: | PROTEIN (FIBRONECTIN) | | Authors: | Potts, J.R, Bright, J.R, Bolton, D, Pickford, A.R, Campbell, I.D. | | Deposit date: | 1999-04-21 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal F1 module pair from human fibronectin.
Biochemistry, 38, 1999
|
|
4H31
 
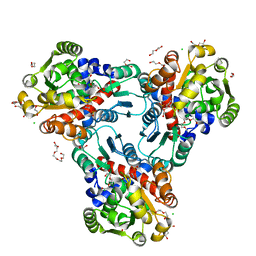 | | Crystal structure of anabolic ornithine carbamoyltransferase from Vibrio vulnificus in complex with carbamoyl phosphate and L-norvaline | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Winsor, J, Grimshaw, S, Osinski, T, Chordia, M.D, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of ornithine carbamoyltransferase from various pathogens
To be Published
|
|
6M48
 
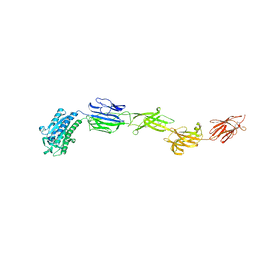 | | Crystal structure of pilus adhesin, SpaC from Lactobacillus rhamnosus GG - P21212 form | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SpaC | | Authors: | Kant, A, Palva, A, Von Ossowaski, I, Krishnan, V. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
5LSI
 
 | | CRYSTAL STRUCTURE OF THE KINETOCHORE MIS12 COMPLEX HEAD2 SUBDOMAIN CONTAINING DSN1 AND NSL1 FRAGMENTS | | Descriptor: | Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, SULFATE ION | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
4KWX
 
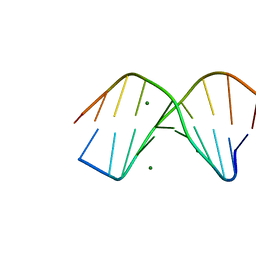 | |
1QZ6
 
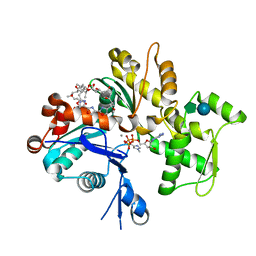 | | Structure of rabbit actin in complex with jaspisamide A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Klenchin, V.A, Allingham, J.S, King, R, Tanaka, J, Marriott, G, Rayment, I. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trisoxazole macrolide toxins mimic the binding of actin-capping proteins to actin
Nat.Struct.Biol., 10, 2003
|
|
4KZB
 
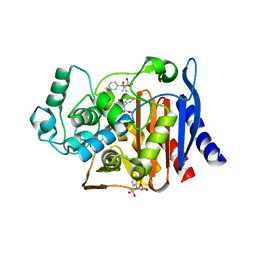 | | Crystal structure of AmpC beta-lactamase in complex with fragment 50 (N-(methylsulfonyl)-N-phenyl-alanine) | | Descriptor: | Beta-lactamase, N-(methylsulfonyl)-N-phenyl-D-alanine, N-(methylsulfonyl)-N-phenyl-L-alanine | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZV
 
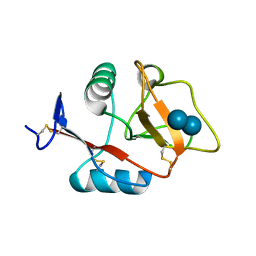 | | Structure of the carbohydrate-recognition domain of the C-type lectin mincle bound to trehalose | | Descriptor: | C-type lectin mincle, CALCIUM ION, SODIUM ION, ... | | Authors: | Feinberg, H, Jegouzo, S.A.F, Rowntree, T.J.W, Guan, Y, Brash, M.A, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2013-05-30 | | Release date: | 2013-08-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism for Recognition of an Unusual Mycobacterial Glycolipid by the Macrophage Receptor Mincle.
J.Biol.Chem., 288, 2013
|
|
4KZ2
 
 | | Crystal Structure of phi29 pRNA 3WJ Core | | Descriptor: | MANGANESE (II) ION, phi29 pRNA 3WJ core RNA 16 mer, phi29 pRNA 3WJ core RNA 18 mer, ... | | Authors: | Zhang, H, Endrizzi, J.A, Shu, Y, Haque, F, Sauter, C, Guo, P, Chi, Y.-I. | | Deposit date: | 2013-05-29 | | Release date: | 2013-10-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of 3WJ core revealing divalent ion-promoted thermostability and assembly of the Phi29 hexameric motor pRNA.
Rna, 19, 2013
|
|
5LUR
 
 | | An avidin mutant | | Descriptor: | Avidin, CHLORIDE ION, PROGESTERONE | | Authors: | Agrawal, N, Lehtonen, S.I, Riihimaki, T.A, Kulomaa, M.S, Hytonen, V.P, Johnson, M.S, Airenne, T.T. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An avidin mutant
TO BE PUBLISHED
|
|
4KZ8
 
 | | Crystal structure of AmpC beta-lactamase in complex with fragment 20 (1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione) | | Descriptor: | 1,3-diethyl-2-thioxodihydropyrimidine-4,6(1H,5H)-dione, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
6HGC
 
 | | Structure of Calypso in complex with DEUBAD of ASX | | Descriptor: | Polycomb protein Asx, Ubiquitin carboxyl-terminal hydrolase calypso,Ubiquitin carboxyl-terminal hydrolase calypso | | Authors: | De, I, Chittock, E.C, Groetsch, H, Miller, T.C.R, McCarthy, A.A, Mueller, C.W. | | Deposit date: | 2018-08-23 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural Basis for the Activation of the Deubiquitinase Calypso by the Polycomb Protein ASX.
Structure, 27, 2019
|
|
3GZD
 
 | | Human selenocysteine lyase, P1 crystal form | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, NITRATE ION, Selenocysteine lyase | | Authors: | Karlberg, T, Hogbom, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
4GXX
 
 | | Crystal structure of the "avianized" 1918 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Ekiert, D.C, Wilson, I.A. | | Deposit date: | 2012-09-04 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Influenza Human Monoclonal Antibody 1F1 Interacts with Three Major Antigenic Sites and Residues Mediating Human Receptor Specificity in H1N1 Viruses.
Plos Pathog., 8, 2012
|
|
6HQ3
 
 | |
4KB9
 
 | | Crystal structure of wild-type HIV-1 protease with novel tricyclic P2-ligands GRL-0739A | | Descriptor: | (3aR,3bR,4S,7aR,8aS)-decahydrofuro[2,3-b][1]benzofuran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2013-04-23 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Highly Potent HIV-1 Protease Inhibitors with Novel Tricyclic P2 Ligands: Design, Synthesis, and Protein-Ligand X-ray Studies.
J.Med.Chem., 56, 2013
|
|
6HQ4
 
 | | Structure of EAL enzyme Bd1971 - cAMP bound form | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, EAL Enzyme Bd1971, MAGNESIUM ION | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2018-09-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Nucleotide signaling pathway convergence in a cAMP-sensing bacterial c-di-GMP phosphodiesterase.
Embo J., 38, 2019
|
|
6HQJ
 
 | |
2ZVN
 
 | | NEMO CoZi domain incomplex with diubiquitin in P212121 space group | | Descriptor: | NF-kappa-B essential modulator, UBC protein | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
8B8F
 
 | | Atomic structure of the beta-trefoil domain of the Laccaria bicolor lectin LBL in complex with lactose | | Descriptor: | N-terminal beta-trefoil domain of the lectin LBL from Laccaria bicolor, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Acebron, I, Campanero-Rhodes, M.A, Solis, D, Menendez, M, Garcia, C, Lillo, M.P, Mancheno, J.M. | | Deposit date: | 2022-10-04 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic crystal structure and sugar specificity of a beta-trefoil lectin domain from the ectomycorrhizal basidiomycete Laccaria bicolor.
Int.J.Biol.Macromol., 233, 2023
|
|
