6MRD
 
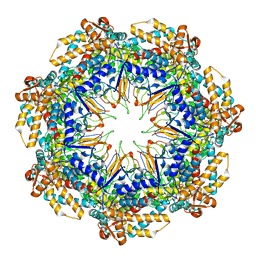 | | ADP-bound human mitochondrial Hsp60-Hsp10 half-football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
2CBI
 
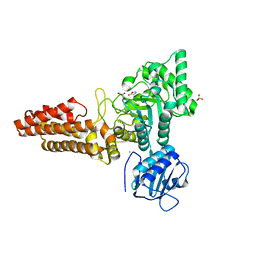 | | Structure of the Clostridium perfringens NagJ family 84 glycoside hydrolase, a homologue of human O-GlcNAcase | | Descriptor: | CHLORIDE ION, GAMMA-BUTYROLACTONE, GLYCEROL, ... | | Authors: | Rao, F.V, Dorfmueller, H.C, Villa, F, Allwood, M, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2006-01-04 | | Release date: | 2006-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis.
EMBO J., 25, 2006
|
|
7YWS
 
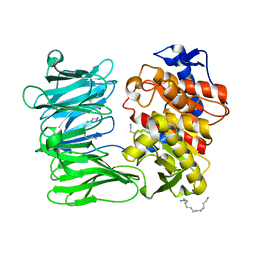 | | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 3 spermine molecules at 1.7 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Elucidation of the Conformational Transition of Oligopeptidase B by an Integrative Approach Based on the Combination of X-ray, SAXS, and Essential Dynamics Sampling Simulation
Crystals, 12, 2022
|
|
6ASU
 
 | |
7YMG
 
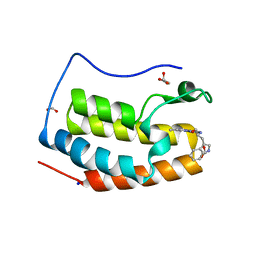 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 2-({3-ethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl}amino)-3-(1H-indol-3-yl)propan-1-ol | | Descriptor: | (2S)-2-[(3-ethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)amino]-3-(1H-indol-3-yl)propan-1-ol, Bromodomain-containing protein 4, FORMIC ACID, ... | | Authors: | Kim, J.H, Lee, B.I. | | Deposit date: | 2022-07-28 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
7YQ9
 
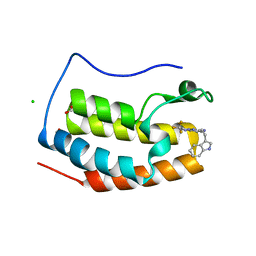 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with N-[2-(1H-indol-3-yl)ethyl]-3-(trifluoromethyl)[1,2,4]triazolo[4,3-b]pyridazin-6-amine | | Descriptor: | Bromodomain-containing protein 4, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, J.H, Lee, B.I. | | Deposit date: | 2022-08-05 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
7YX7
 
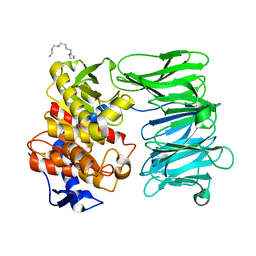 | | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 1 spermine molecule at 1.72 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-15 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Elucidation of the Conformational Transition of Oligopeptidase B by an Integrative Approach Based on the Combination of X-ray, SAXS, and Essential Dynamics Sampling Simulation
Crystals, 12, 2022
|
|
1EC9
 
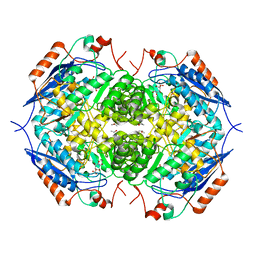 | | E. COLI GLUCARATE DEHYDRATASE BOUND TO XYLAROHYDROXAMATE | | Descriptor: | GLUCARATE DEHYDRATASE, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Gulick, A.M, Hubbard, B.K, Gerlt, J.A, Rayment, I. | | Deposit date: | 2000-01-25 | | Release date: | 2000-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystallographic and mutagenesis studies of the reaction catalyzed by D-glucarate dehydratase from Escherichia coli.
Biochemistry, 39, 2000
|
|
6AWA
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-05 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Pseudomonas putida in Complex with FAD and Adenosine-5'-monophosphate.
To Be Published
|
|
6N8Q
 
 | |
1NB5
 
 | | Crystal structure of stefin A in complex with cathepsin H | | Descriptor: | Cathepsin H, Cathepsin H MINI CHAIN, STEFIN A, ... | | Authors: | Jenko, S, Dolenc, I, Guncar, G, Dobersek, A, Podobnik, M, Turk, D. | | Deposit date: | 2002-12-02 | | Release date: | 2003-02-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases
J.Mol.Biol., 326, 2003
|
|
5VJM
 
 | | Crystal structure of H7 hemagglutinin mutant (V186K, K193T, G228S) from the influenza virus A/Shanghai/2/2013 (H7N9) with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, Hemagglutinin HA2, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2017-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Three mutations switch H7N9 influenza to human-type receptor specificity.
PLoS Pathog., 13, 2017
|
|
3E0J
 
 | | X-ray structure of the complex of regulatory subunits of human DNA polymerase delta | | Descriptor: | DNA polymerase subunit delta-2, DNA polymerase subunit delta-3 | | Authors: | Baranovskiy, A.G, Babayeva, N.D, Pavlov, Y.I, Vassylyev, D.G, Tahirov, T.H. | | Deposit date: | 2008-07-31 | | Release date: | 2008-12-16 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of the complex of regulatory subunits of human DNA polymerase delta.
Cell Cycle, 7, 2008
|
|
7KO6
 
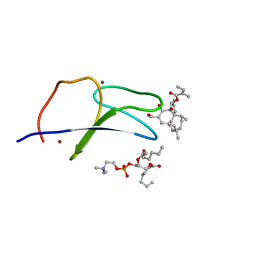 | | C1B domain of Protein kinase C in complex with ingenol-3-angelate and phosphocholine | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, Protein kinase C delta type, ZINC ION, ... | | Authors: | Katti, S.S, Krieger, I.V. | | Deposit date: | 2020-11-06 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural anatomy of Protein Kinase C C1 domain interactions with diacylglycerol and other agonists.
Nat Commun, 13, 2022
|
|
2JSB
 
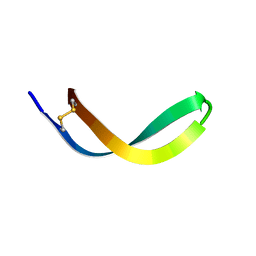 | | Solution structure of arenicin-1 | | Descriptor: | Arenicin-1 | | Authors: | Jakovkin, I.B, Hecht, O, Gelhaus, C, Krasnosdembskaya, A.D, Fedders, H, Leippe, M, Groetzinger, J. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-05 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Structure and mode of action of the antimicrobial peptide arenicin
Biochem.J., 410, 2008
|
|
5BRL
 
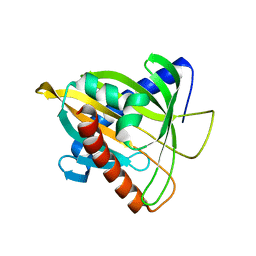 | | Crystal Structure of L124D STARD4 | | Descriptor: | StAR-related lipid transfer protein 4 | | Authors: | Iaea, D.B, Dikiy, I, Kiburu, I, Eliezer, D.E, Maxfield, F.R. | | Deposit date: | 2015-05-31 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | STARD4 Membrane Interactions and Sterol Binding.
Biochemistry, 54, 2015
|
|
6AS6
 
 | |
4YEM
 
 | | Carboplatin binding to HEWL in NaBr crystallisation conditions studied at an X-ray wavelength of 0.9163A - new refinement | | Descriptor: | ACETATE ION, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Shabalin, I.G, Dauter, Z, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallography and chemistry should always go together: a cautionary tale of protein complexes with cisplatin and carboplatin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2JVZ
 
 | | Solution NMR Structure of the Second and Third KH Domains of KSRP | | Descriptor: | Far upstream element-binding protein 2 | | Authors: | Diaz-Moreno, I, Hollingworth, D, Garcia-Mayoral, M.F, Kelly, G, Cukier, C.D, Ramos, A. | | Deposit date: | 2007-09-28 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Second and Third KH Domains of KSRP
To be Published, 2007
|
|
7YWZ
 
 | | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 4 spermine molecules at 1.75 A resolution | | Descriptor: | GLYCEROL, Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 4 spermine molecules at 1.75 A resolution
To Be Published
|
|
1G2I
 
 | | CRYSTAL STRUCTURE OF A NOVEL INTRACELLULAR PROTEASE FROM PYROCOCCUS HORIKOSHII AT 2 A RESOLUTION | | Descriptor: | PROTEASE I, SULFATE ION | | Authors: | Du, X, Choi, I.-G, Kim, R, Jancarik, J, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-10-19 | | Release date: | 2000-11-08 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-A resolution.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1NQU
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 6,7-dioxo-5H-8-ribitylaminolumazine | | Descriptor: | 6,7-DIOXO-5H-8-RIBITYLAMINOLUMAZINE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
4OEH
 
 | | X-ray Structure of Uridine Phosphorylase from Vibrio cholerae Complexed with Uracil at 1.91 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases.
To Be Published, Crystallogr. Rep., 2016
|
|
6L8M
 
 | | WNT DNA promoter mutant G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*CP*AP*CP*CP*GP*GP*GP*CP*AP*GP*TP*GP*GP*GP*CP*GP*GP*G)-3') | | Authors: | Wang, Z.F, Li, M.H, Chu, I.T, Winnerdy, F.R, Phan, A.T, Chang, T.C. | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cytosine epigenetic modification modulates the formation of an unprecedented G4 structure in the WNT1 promoter.
Nucleic Acids Res., 48, 2020
|
|
5VFG
 
 | |
