2PPC
 
 | |
6FCZ
 
 | | Model of gC1q-Fc complex based on 7A EM map | | Descriptor: | Complement C1q subcomponent subunit A, Complement C1q subcomponent subunit B, Complement C1q subcomponent subunit C, ... | | Authors: | Ugurlar, D, Howes, S.C, de Kreuk, B.J.K, de Jong, R.N, Beurskens, F.J, Koster, A.J, Parren, P.W.H.I, Sharp, T.H, Gros, P, Koning, R.I. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-28 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement.
Science, 359, 2018
|
|
1I7M
 
 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COMPLEXED WITH 4-AMIDINOINDAN-1-ONE-2'-AMIDINOHYDRAZONE | | Descriptor: | 1,4-DIAMINOBUTANE, 4-AMIDINOINDAN-1-ONE-2'-AMIDINOHYDRAZONE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-09 | | Release date: | 2001-08-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
5KJ7
 
 | | Structure of the Ca2+-bound synaptotagmin-1 SNARE complex (long unit cell form) - from XFEL diffraction | | Descriptor: | CALCIUM ION, Synaptosomal-associated protein 25, Synaptotagmin-1, ... | | Authors: | Lyubimov, A.Y, Uervirojnangkoorn, M, Zhou, Q, Zhao, M, Sauter, N.K, Brewster, A.S, Weis, W.I, Brunger, A.T. | | Deposit date: | 2016-06-17 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Advances in X-ray free electron laser (XFEL) diffraction data processing applied to the crystal structure of the synaptotagmin-1 / SNARE complex.
Elife, 5, 2016
|
|
1WLS
 
 | | Crystal structure of L-asparaginase I homologue protein from Pyrococcus horikoshii | | Descriptor: | L-asparaginase | | Authors: | Yao, M, Morita, H, Yasutake, Y, Tanaka, I. | | Deposit date: | 2004-06-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of the type I L-asparaginase from the hyperthermophilic archaeon Pyrococcus horikoshii at 2.16 angstroms resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4YPE
 
 | | ASH1L SET domain H2193F mutant in complex with S-adenosyl methionine (SAM) | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Rogawski, D.S, Ndoj, J, Cho, H.J, Maillard, I, Grembecka, J, Cierpicki, T. | | Deposit date: | 2015-03-12 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase.
Biochemistry, 54, 2015
|
|
4YNM
 
 | | ASH1L wild-type SET domain in complex with S-adenosyl methionine (SAM) | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Rogawski, D.S, Ndoj, J, Cho, H.-J, Maillard, I, Grembecka, J, Cierpicki, T. | | Deposit date: | 2015-03-10 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase.
Biochemistry, 54, 2015
|
|
8QU3
 
 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
8R1D
 
 | | SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-3 Fab Heavy Chain, SD1-3 Fab Light Chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8QU4
 
 | | NF-YB/C Heterodimer in Complex with a 13-mer NF-YA-derived Peptide Stabilized with C8-Hydrocarbon Linker in an alternative binding pose | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Nuclear transcription factor Y subunit alpha, ... | | Authors: | Arbore, F, Durukan, C, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2023-10-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Binding Dynamics of a Stapled Peptide Targeting the Transcription Factor NF-Y.
Chembiochem, 25, 2024
|
|
6URS
 
 | | Sleeping Beauty transposase PAI subdomain mutant - H19Y | | Descriptor: | Sleeping Beauty transposase PAI subdomain | | Authors: | Nesmelova, I.V, Leighton, G.O, Yan, C, Lustig, J, Corona, R.I, Guo, J.T, Ivics, Z. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | H19Y mutation in the primary DNA-recognition subdomain of the Sleeping Beauty transposase improves structural stability, transposon DNA-binding and transposition
To Be Published
|
|
6WO5
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 1a bound to neutralizing antibody 212.1.1 and non neutralizing antibody E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-24 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
8S2X
 
 | |
6IOQ
 
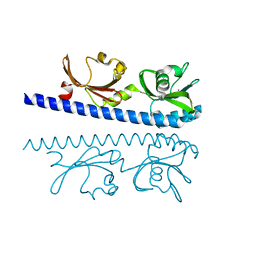 | | The ligand binding domain of Mlp24 with glycine | | Descriptor: | CALCIUM ION, GLYCINE, Methyl-accepting chemotaxis protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
8S2W
 
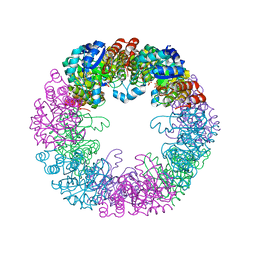 | |
6SP1
 
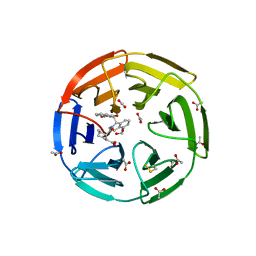 | | KEAP1 IN COMPLEX WITH COMPOUND 6 | | Descriptor: | (1~{S},2~{R})-2-[[(1~{S})-1-[[1,3-bis(oxidanylidene)isoindol-2-yl]methyl]-5-(2-hydroxyethyloxy)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]cyclohexane-1-carboxylic acid, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Ontoria, J.M, Biancofiore, I, Fezzardi, P, Torrente de Haro, E, Colarusso, S, Bianchi, E, Andreini, M, Patsilinakos, A, Summa, V, Pacifici, R, Munoz-Sanjuan, I, Park, L, Bresciani, A, Dominguez, C, Toledo-Sherman, L, Harper, S. | | Deposit date: | 2019-08-30 | | Release date: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Combined Peptide and Small-Molecule Approach toward Nonacidic THIQ Inhibitors of the KEAP1/NRF2 Interaction.
Acs Med.Chem.Lett., 11, 2020
|
|
8SIT
 
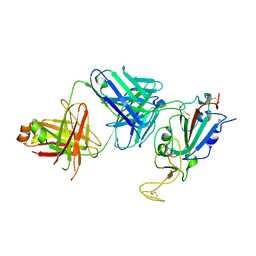 | |
1V49
 
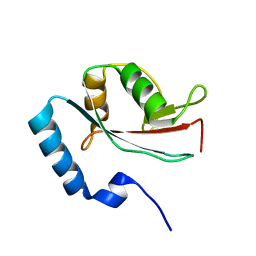 | | Solution structure of microtubule-associated protein light chain-3 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kouno, T, Mizuguchi, M, Tanida, I, Ueno, T, Kominami, E, Kawano, K. | | Deposit date: | 2003-11-11 | | Release date: | 2004-12-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains.
J.Biol.Chem., 280, 2005
|
|
8ROX
 
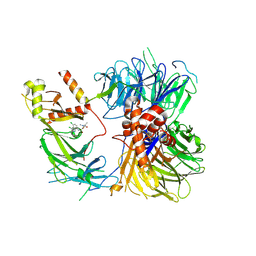 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | Descriptor: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
8ROY
 
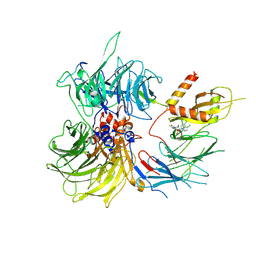 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | Descriptor: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
6FPJ
 
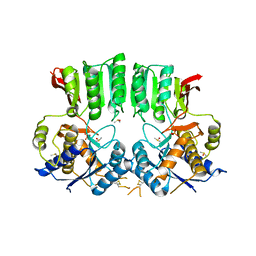 | | Structure of the AMPAR GluA3 N-terminal domain bound to phosphate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Garcia-Nafria, J, Greger, I. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain.
Structure, 27, 2019
|
|
4YPU
 
 | | ASH1L SET domain K2264L mutant in complex with S-adenosyl methionine (SAM) | | Descriptor: | Histone-lysine N-methyltransferase ASH1L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Rogawski, D.S, Ndoj, J, Cho, H.J, Maillard, I, Grembecka, J, Cierpicki, T. | | Deposit date: | 2015-03-13 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Two Loops Undergoing Concerted Dynamics Regulate the Activity of the ASH1L Histone Methyltransferase.
Biochemistry, 54, 2015
|
|
1EBE
 
 | | Laue diffraction study on the structure of cytochrome c peroxidase compound I | | Descriptor: | CYTOCHROME C PEROXIDASE, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fulop, V, Phizackerley, R.P, Soltis, S.M, Clifton, I.J, Wakatsuki, S, Erman, J.E, Hajdu, J, Edwards, S.L. | | Deposit date: | 2001-07-25 | | Release date: | 2001-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Laue Diffraction Study on the Structure of Cytochrome C Peroxidase Compound I
Structure, 2, 1994
|
|
6R70
 
 | | Endogeneous native human 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Schmidli, C, Albiez, S, Rima, L, Righetto, R, Mohammed, I, Oliva, P, Kovacik, L, Stahlberg, H, Braun, T. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Microfluidic protein isolation and sample preparation for high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4YTB
 
 | | Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) in complex with dipeptide Asp-Gln. | | Descriptor: | ASPARTIC ACID, AZIDE ION, CHLORIDE ION, ... | | Authors: | Goulas, T, Mizgalska, D, Garcia-Ferrer, I, Kantyka, T, Guevara, T, Szmigielski, B, Sroka, A, Millan, C, Uson, I, Veillard, F, Potempa, B, Mydel, P, Sola, M, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-17 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase.
Sci Rep, 5, 2015
|
|
