5NZM
 
 | | Crystal structure of UDP-glucose pyrophosphorylase from Leishmania major in complex with murrayamine-I | | Descriptor: | Murrayamine-I, UDP-glucose pyrophosphorylase | | Authors: | Cramer, J.T, Fuehring, J.I, Baruch, P, Bruetting, C, Hesse, R, Knoelker, H.-J, Gerardy-Schahn, R, Fedorov, R. | | Deposit date: | 2017-05-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies
Acs Catalysis, 8, 2018
|
|
6XPP
 
 | |
6XOA
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with C145S mutation | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with C145S mutation
To Be Published
|
|
6X8O
 
 | | BimBH3 peptide tetramer | | Descriptor: | Bcl-2-like protein 11, THIOCYANATE ION | | Authors: | Robin, A.Y, Westphal, D, Uson, I, Czabotar, P.E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-09-09 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Biophysical Characterization of Pro-apoptotic BimBH3 Peptides Reveals an Unexpected Capacity for Self-Association.
Structure, 29, 2021
|
|
4GHA
 
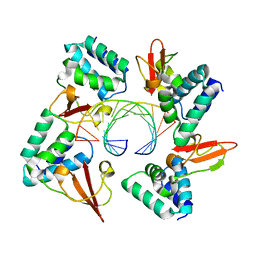 | | Crystal structure of Marburg virus VP35 RNA binding domain bound to 12-bp dsRNA | | Descriptor: | Polymerase cofactor VP35, RNA (5'-R(*CP*UP*AP*GP*AP*CP*GP*UP*CP*UP*AP*G)-3') | | Authors: | Bale, S, Jean-Philippe, J, Bornholdt, Z.A, Kimberlin, C.K, Halfmann, P, Zandonatti, M.A, Kunert, J, Kroon, G.J.A, Kawaoka, Y, MacRae, I.J, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Marburg Virus VP35 Can Both Fully Coat the Backbone and Cap the Ends of dsRNA for Interferon Antagonism.
Plos Pathog., 8, 2012
|
|
7QUH
 
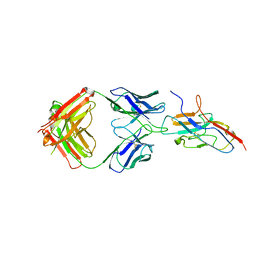 | | Siglec-8 in complex with therapeutic Fab AK002. | | Descriptor: | Sialic acid-binding Ig-like lectin 8, Sialic acid-binding immunoglobulin-type lectin | | Authors: | Lenza, M.P, Oyenarte, I, Jimenez Barbero, J, Ereno Orbea, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
5L3Q
 
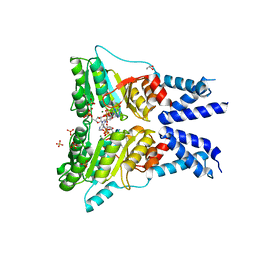 | | Structure of the GTPase heterodimer of human SRP54 and SRalpha | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wild, K, Segnitz, B, Sinning, I. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for Conserved Regulation and Adaptation of the Signal Recognition Particle Targeting Complex.
J.Mol.Biol., 428, 2016
|
|
2MSB
 
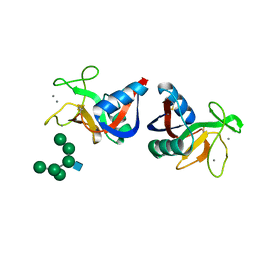 | | STRUCTURE OF A C-TYPE MANNOSE-BINDING PROTEIN COMPLEXED WITH AN OLIGOSACCHARIDE | | Descriptor: | CALCIUM ION, MANNOSE-BINDING PROTEIN-A, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Weis, W.I, Drickamer, K, Hendrickson, W.A. | | Deposit date: | 1992-07-28 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a C-type mannose-binding protein complexed with an oligosaccharide.
Nature, 360, 1992
|
|
6XU1
 
 | | Crystal structure of tetrameric human H215A-SAMHD1 (residues 109-626) with GTP, dAMPNPP and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
8AIP
 
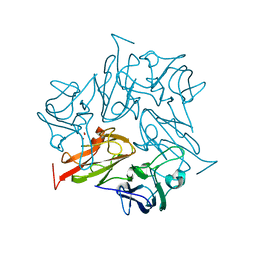 | | Crystal Structure of Two-domain bacterial laccase from the actinobacterium Streptomyces carpinensis VKM Ac-1300 | | Descriptor: | COPPER (II) ION, OXYGEN MOLECULE, Two-Domain Laccase | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Trubitsina, L, Trubitsin, I, Leontievsky, A, Lisov, A. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Novel Two-Domain Laccase with Middle Redox Potential: Physicochemical and Structural Properties.
Biochemistry Mosc., 88, 2023
|
|
8AF1
 
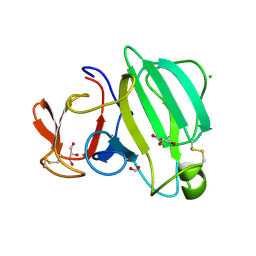 | | Beta-Lytic Protease from Lysobacter capsici | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kudryakova, I.V, Afoshin, A.S, Vasilyeva, N.V. | | Deposit date: | 2022-07-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural and Functional Characterization of beta-lytic Protease from Lysobacter capsici VKM B-2533 T.
Int J Mol Sci, 23, 2022
|
|
1H3F
 
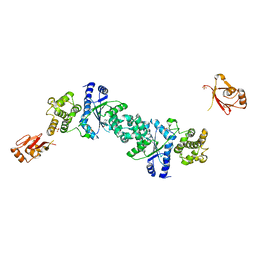 | | Tyrosyl-tRNA synthetase from Thermus thermophilus complexed with tyrosinol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, SULFATE ION, TYROSYL-TRNA SYNTHETASE | | Authors: | Cusack, S, Yaremchuk, A, Kriklivyi, I, Tukalo, M. | | Deposit date: | 2002-08-28 | | Release date: | 2002-09-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Class I Tyrosyl-tRNA Synthetase Has a Class II Mode or tRNA Recognition
Embo J., 21, 2002
|
|
6XQE
 
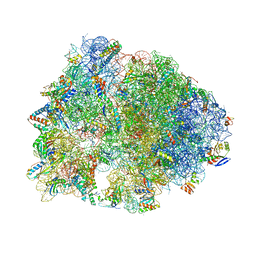 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with sarecycline, UAA-mRNA, and deacylated P-site tRNA at 3.00A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Lomakin, I.B, Bunick, C.G, Polikanov, Y.S. | | Deposit date: | 2020-07-09 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Sarecycline interferes with tRNA accommodation and tethers mRNA to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7RNJ
 
 | | S2P6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | Monoclonal antibody S2P6 Fab heavy chain, Monoclonal antibody S2P6 Fab light chain, SULFATE ION, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M, Sauer, M.M, Veesler, D. | | Deposit date: | 2021-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Broad betacoronavirus neutralization by a stem helix-specific human antibody.
Science, 373, 2021
|
|
8OJ8
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 1 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
6CHT
 
 | | HNF4alpha in complex with the corepressor EBP1 fragment | | Descriptor: | Hepatocyte nuclear factor 4-alpha, LAURIC ACID, Proliferation-associated protein 2G4 | | Authors: | Chi, Y.I, Singh, P, Lee, I.K. | | Deposit date: | 2018-02-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.174 Å) | | Cite: | ErbB3-binding protein 1 (EBP1) represses HNF4 alpha-mediated transcription and insulin secretion in pancreatic beta-cells.
J.Biol.Chem., 294, 2019
|
|
5EDL
 
 | | Crystal structure of an S-component of ECF transporter | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Putative HMP/thiamine permease protein YkoE, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Josts, I, Tidow, H. | | Deposit date: | 2015-10-21 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a Group I Energy Coupling Factor Vitamin Transporter S Component in Complex with Its Cognate Substrate.
Cell Chem Biol, 23, 2016
|
|
8AKU
 
 | | 250 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKY
 
 | | 290 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKZ
 
 | | 320 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AL0
 
 | | 365 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKX
 
 | | 305 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKV
 
 | | 270 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKW
 
 | | 280 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
1DTT
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 (PETT130A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-04-02 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
