6ZMZ
 
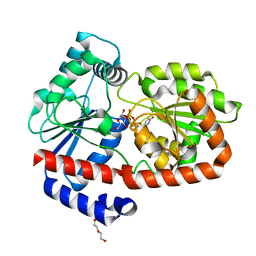 | | UDPG-bound Trehalose transferase from Thermoproteus uzoniensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Trehalose phosphorylase/synthase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | Deposit date: | 2020-07-05 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
Acs Catalysis, 10, 2020
|
|
4TNN
 
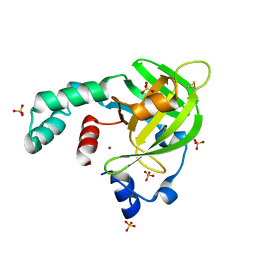 | | Crystal structure of Escherichia coli protein YodA in complex with Ni - artifact of purification. | | Descriptor: | Metal-binding lipocalin, NICKEL (II) ION, SULFATE ION | | Authors: | Gasiorowska, O.A, Cymborowski, M.T, Handing, K.B, Shabalin, I.G, Zasadzinska, E, Niedzialkowska, E, Porebski, P.J, Minor, W. | | Deposit date: | 2014-06-04 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
6ZQJ
 
 | | Cryo-EM structure of trimeric prME spike of Spondweni virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, prM | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-09 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
1RK1
 
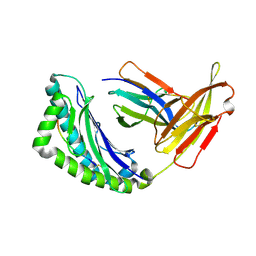 | | Mhc Class I Natural H-2Kb Heavy Chain Complexed With beta-2 Microglobulin and Herpes Simplex Virus Mutant Glycoprotein B Peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Glycoprotein B, ... | | Authors: | Miley, M.J, Messaoudi, I, Nikolich-Zugich, J, Fremont, D.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Restoration of TCR Recognition of an MHC Allelic Variant by Peptide Secondary Anchor Substitution
J.Exp.Med., 200, 2004
|
|
6O9G
 
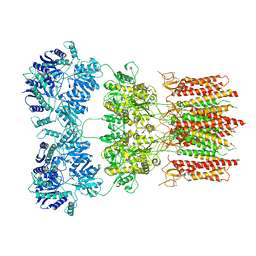 | | Open state GluA2 in complex with STZ and blocked by AgTx-636, after micelle signal subtraction | | Descriptor: | CYCLOTHIAZIDE, GLUTAMIC ACID, Glutamate receptor 2,Voltage-dependent calcium channel gamma-2 subunit, ... | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Vassilevski, A.A, Sobolevsky, A.I. | | Deposit date: | 2019-03-13 | | Release date: | 2019-03-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors.
Neuron, 99, 2018
|
|
6O5X
 
 | | Crystal Structure of multi-drug resistant HIV-1 protease PR-S17 with substrate analog CA-p2 | | Descriptor: | CHLORIDE ION, HIV-1 protease PR-S17, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, ... | | Authors: | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2019-03-04 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Highly Drug-Resistant HIV-1 Protease Mutant PRS17 Shows Enhanced Binding to Substrate Analogues.
Acs Omega, 4, 2019
|
|
8AKQ
 
 | | 180 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKR
 
 | | 200 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKT
 
 | | 235 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKS
 
 | | 215 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
6OJ1
 
 | | Crystal Structure of Aspergillus fumigatus Ega3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bamford, N.C, Subramanian, A.S, Millan, C, Uson, I, Howell, P.L. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Ega3 from the fungal pathogenAspergillus fumigatusis an endo-alpha-1,4-galactosaminidase that disrupts microbial biofilms.
J.Biol.Chem., 294, 2019
|
|
8G2B
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAphe, and aminoacylated P-site fMet-tRNAmet at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
8G2A
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-tRNAphe, peptidyl P-site fMTHSMRC-tRNAmet, and deacylated E-site tRNAphe at 2.45A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
6OMB
 
 | | Cdc48 Hexamer (Subunits A to E) with substrate bound to the central pore | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division control protein 48, ... | | Authors: | Cooney, I, Han, H, Stewart, M, Carson, R.H, Hansen, D, Price, J.C, Hill, C.P, Shen, P.S. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Cdc48 segregase in the act of unfolding an authentic substrate.
Science, 365, 2019
|
|
2DB4
 
 | | Crystal structure of rotor ring with DCCD of the V- ATPase from Enterococcus hirae | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DICYCLOHEXYLUREA, SODIUM ION, ... | | Authors: | Murata, T, Yamato, I, Kakinuma, Y, Shirouzu, M, Walker, J.E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-05 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the rotor ring modified with N,N'-dicyclohexylcarbodiimide of the Na+-transporting vacuolar ATPase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5NCW
 
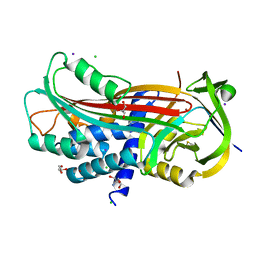 | | Structure of the trypsin induced serpin-type proteinase inhibitor, miropin (V367K/K368A mutant). | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goulas, T, Ksiazek, M, Garcia-Ferrer, I, Mizgalska, D, Potempa, J, Gomis-Ruth, X. | | Deposit date: | 2017-03-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A structure-derived snap-trap mechanism of a multispecific serpin from the dysbiotic human oral microbiome.
J. Biol. Chem., 292, 2017
|
|
4RNY
 
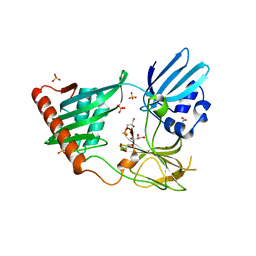 | | Structure of Helicobacter pylori Csd3 from the orthorhombic crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, SULFATE ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6ZVK
 
 | | The Halastavi arva virus (HalV) intergenic region IRES promotes translation by the simplest possible initiation mechanism | | Descriptor: | 18S RIBOSOMAL RNA, 28S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN ES17, ... | | Authors: | Abaeva, I.S, Vicens, Q, Bochler, A, Soufari, H, Simonetti, A, Pestova, T, Hashem, Y, Hellen, C.U.T. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The Halastavi arva Virus Intergenic Region IRES Promotes Translation by the Simplest Possible Initiation Mechanism.
Cell Rep, 33, 2020
|
|
8T3P
 
 | |
5E79
 
 | | Macromolecular diffractive imaging using imperfect crystals | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthur, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffer, A, Dorner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
3JTM
 
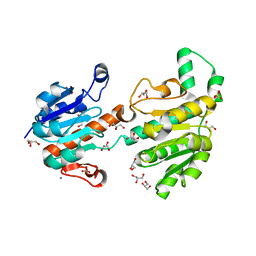 | | Structure of recombinant formate dehydrogenase from Arabidopsis thaliana | | Descriptor: | AZIDE ION, Formate dehydrogenase, mitochondrial, ... | | Authors: | Timofeev, V.I, Shabalin, I.G, Serov, A.E, Polyakov, K.M, Popov, V.O, Tishkov, V.I, Kuranova, I.P, Samigina, V.R. | | Deposit date: | 2009-09-13 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of recombinant formate dehydrogenase from Arabidopsis thaliana
to be published
|
|
8A1F
 
 | | Human PTPRK N-terminal domains MAM-Ig-FN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hay, I.M, Graham, S.C, Sharpe, H.J, Deane, J.E. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains.
J.Biol.Chem., 299, 2023
|
|
8VC8
 
 | |
7UFI
 
 | | VchTnsC AAA+ ATPase with DNA, single heptamer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*TP*CP*CP*AP*GP*TP*AP*CP*AP*GP*CP*GP*CP*GP*GP*CP*TP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*AP*GP*CP*CP*GP*CP*GP*CP*TP*GP*TP*AP*CP*TP*GP*GP*AP*G)-3'), ... | | Authors: | Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Selective TnsC recruitment enhances the fidelity of RNA-guided transposition.
Nature, 609, 2022
|
|
