1ZA3
 
 | | The crystal structure of the YSd1 Fab bound to DR5 | | Descriptor: | Fab-YSd1 heavy chain, Fab-YSd1 light chain, Tumor necrosis factor receptor superfamily member 10B | | Authors: | Fellouse, F.A, Li, B, Compaan, D.M, Peden, A.A, Hymowitz, S.G, Sidhu, S.S. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Molecular recognition by a binary code.
J.Mol.Biol., 348, 2005
|
|
3NOB
 
 | |
3OJ4
 
 | |
3OJ3
 
 | |
3P08
 
 | |
3PTF
 
 | |
3UW5
 
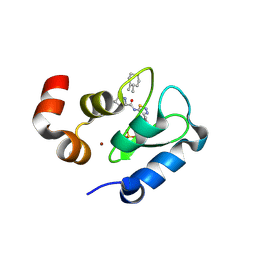 | | Crystal structure of the BIR domain of MLIAP bound to GDC0152 | | Descriptor: | Baculoviral IAP repeat-containing protein 7, Baculoviral IAP repeat-containing protein 4, GDC-0152, ... | | Authors: | Maurer, B, Hymowitz, S.G. | | Deposit date: | 2011-11-30 | | Release date: | 2012-02-22 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of a Potent Small-Molecule Antagonist of Inhibitor of Apoptosis (IAP) Proteins and Clinical Candidate for the Treatment of Cancer (GDC-0152).
J.Med.Chem., 55, 2012
|
|
4MXW
 
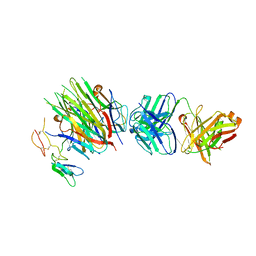 | | Structure of heterotrimeric lymphotoxin LTa1b2 bound to lymphotoxin beta receptor LTbR and anti-LTa Fab | | Descriptor: | Lymphotoxin-alpha, Lymphotoxin-beta, Tumor necrosis factor receptor superfamily member 3, ... | | Authors: | Sudhamsu, J, Yin, J.P, Hymowitz, S.G. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Dimerization of LT beta R by LT alpha 1 beta 2 is necessary and sufficient for signal transduction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3U30
 
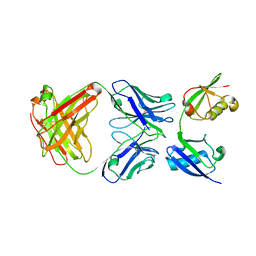 | | Crystal structure of a linear-specific Ubiquitin fab bound to linear ubiquitin | | Descriptor: | Heavy chain Fab, Light chain Fab, linear di-ubiquitin | | Authors: | Matsumoto, M.L, Dong, K.C, Yu, C, Phu, L, Gao, X, Hannoush, R.N, Hymowitz, S.G, Kirkpatrick, D.S, Dixit, V.M, Kelley, R.F. | | Deposit date: | 2011-10-04 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Engineering and structural characterization of a linear polyubiquitin-specific antibody.
J.Mol.Biol., 418, 2012
|
|
4MXV
 
 | | Structure of Lymphotoxin alpha bound to anti-LTa Fab | | Descriptor: | Lymphotoxin-alpha, anti-Lymphotoxin alpha antibody heavy chain, anti-Lymphotoxin alpha antibody light chain | | Authors: | Yin, J.P, Hymowitz, S.G. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dimerization of LT beta R by LT alpha 1 beta 2 is necessary and sufficient for signal transduction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4QM0
 
 | | Crystal structure of RORc in complex with a tertiary sulfonamide inverse agonist | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-methylpropyl)-N-({5-[4-(methylsulfonyl)phenyl]thiophen-2-yl}methyl)-1-phenylmethanesulfonamide, Nuclear receptor ROR-gamma | | Authors: | Boenig, G, Hymowitz, S.G, Wang, W. | | Deposit date: | 2014-06-14 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Reduction in lipophilicity improved the solubility, plasma-protein binding, and permeability of tertiary sulfonamide RORc inverse agonists.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1OSX
 
 | | Solution Structure of the Extracellular Domain of BLyS Receptor 3 (BR3) | | Descriptor: | Tumor necrosis factor receptor superfamily member 13C | | Authors: | Gordon, N.C, Pan, B, Hymowitz, S.G, Yin, J.P, Kelley, R.F, Cochran, A.G, Yan, M, Dixit, V.M, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-03-20 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Biochemistry, 42, 2003
|
|
1OSG
 
 | | Complex between BAFF and a BR3 derived peptide presented in a beta-hairpin scaffold | | Descriptor: | BR3 derived PEPTIDE, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B | | Authors: | Gordon, N.C, Pan, B, Hymowitz, S.G, Yin, J.P, Kelley, R.F, Cochran, A.G, Yan, M, Dixit, V.M, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-03-19 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Biochemistry, 42, 2003
|
|
5WNM
 
 | |
1U5Z
 
 | | The Crystal structure of murine APRIL, pH 8.5 | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13 | | Authors: | Wallweber, H.J, Compaan, D.M, Starovasnik, M.A, Hymowitz, S.G. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of A Proliferation-inducing Ligand, APRIL.
J.Mol.Biol., 343, 2004
|
|
1U5Y
 
 | | Crystal structure of murine APRIL, pH 8.0 | | Descriptor: | Tumor necrosis factor ligand superfamily member 13 | | Authors: | Wallweber, H.J, Compaan, D.M, Starovasnik, M.A, Hymowitz, S.G. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of A Proliferation-inducing Ligand, APRIL.
J.Mol.Biol., 343, 2004
|
|
1U5X
 
 | | Crystal structure of murine APRIL at pH 5.0 | | Descriptor: | Tumor necrosis factor ligand superfamily member 13 | | Authors: | Wallweber, H.J, Compaan, D.M, Starovasnik, M.A, Hymowitz, S.G. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of A Proliferation-inducing Ligand, APRIL.
J.Mol.Biol., 343, 2004
|
|
5T8O
 
 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) bound to Imidazobenzoxepin Compound 3 | | Descriptor: | 10-(3-methyl-3-oxidanyl-but-1-ynyl)-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 14, SULFATE ION | | Authors: | Smith, M.A, McEwan, P, Hymowitz, S.G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-Based Design of Tricyclic NF-kappa B Inducing Kinase (NIK) Inhibitors That Have High Selectivity over Phosphoinositide-3-kinase (PI3K).
J. Med. Chem., 60, 2017
|
|
5T8Q
 
 | |
