8DL3
 
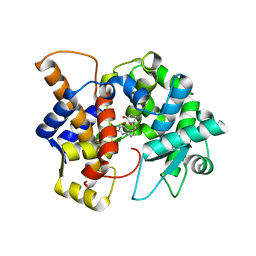 | | Crystal structure of the human queuine salvage enzyme DUF2419, complexed with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, Queuosine salvage protein | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-07-06 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7UI4
 
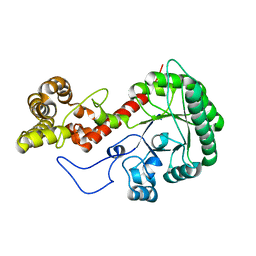 | |
3UD8
 
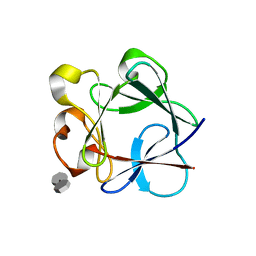 | | Crystal Structure Analysis of FGF1-Disaccharide(NI22) complex | | Descriptor: | 2-deoxy-3-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
3UD9
 
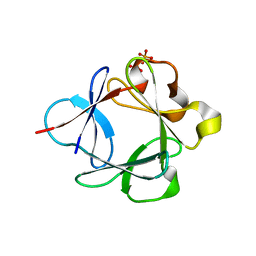 | | Crystal Structure Analysis of FGF1-Disaccharide(NI23) complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
3UDA
 
 | | Crystal Structure Analysis of FGF1-Disaccharide(NI24) complex | | Descriptor: | 2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
7UK3
 
 | |
7UGK
 
 | |
3UD7
 
 | | Crystal Structure Analysis of FGF1-Disaccharide(NI21) complexes | | Descriptor: | 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Heparin-binding growth factor 1, PHOSPHATE ION | | Authors: | Hung, S.-C, Shi, Z. | | Deposit date: | 2011-10-27 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction
J.Am.Chem.Soc., 134, 2012
|
|
7U07
 
 | |
7U1O
 
 | | Crystal structure of queuine salvage enzyme DUF2419 complexed with queuosine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-beta-D-ribofuranosyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7U5A
 
 | | Crystal structure of queuine salvage enzyme DUF2419 mutant K199C, complexed with queuosine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-beta-D-ribofuranosyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, MALONATE ION, Queuine salvage enzyme DUF2419 | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-03-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7U91
 
 | | Crystal structure of queuine salvage enzyme DUF2419, in complex with queuosine-5'-monophosphate | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-(5-O-phosphono-beta-D-ribofuranosyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
7ULC
 
 | | Crystal structure of queuine salvage enzyme DUF2419 mutant D231N, in complex with queuosine-5'-monophosphate | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-7-(5-O-phosphono-beta-D-ribofuranosyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, DI(HYDROXYETHYL)ETHER, Queuosine salvage protein DUF2419 | | Authors: | Hung, S.-H, Swairjo, M.A. | | Deposit date: | 2022-04-04 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of Qng1-mediated salvage of the micronutrient queuine from queuosine-5'-monophosphate as the biological substrate.
Nucleic Acids Res., 51, 2023
|
|
4DCP
 
 | | Crystal Structure of caspase 3, L168F mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
1MQ0
 
 | | Crystal Structure of Human Cytidine Deaminase | | Descriptor: | 1-BETA-RIBOFURANOSYL-1,3-DIAZEPINONE, Cytidine Deaminase, ZINC ION | | Authors: | Chung, S.J, Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-09-13 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human cytidine deaminase bound to a potent inhibitor
J.Med.Chem., 48, 2005
|
|
1M3H
 
 | | Crystal Structure of Hogg1 D268E Mutant with Product Oligonucleotide | | Descriptor: | 5'-D(P*GP*CP*GP*TP*CP*CP*AP*(DDX))-3', 5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*TP*AP*CP*C)-3', ... | | Authors: | Chung, S.J, Verdine, G.L. | | Deposit date: | 2002-06-27 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of End Products Resulting from Lesion Processing by a DNA Glycosylase/Lyase
Chem.Biol., 11, 2004
|
|
1M3Q
 
 | |
4DCO
 
 | | Crystal Structure of caspase 3, L168Y mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-18 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
4DCJ
 
 | | Crystal structure of caspase 3, L168D mutant | | Descriptor: | Caspase Inhibitor AC-DEVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Chung, S.J, Kang, H.J, Kim, S.J. | | Deposit date: | 2012-01-17 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insight into the role of the leucine residue on the L2 loop in the catalytic activity of caspases 3 and 7
Biosci.Rep., 32, 2012
|
|
7WRS
 
 | |
7WRU
 
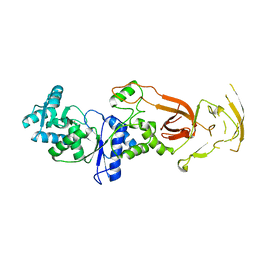 | |
5KUC
 
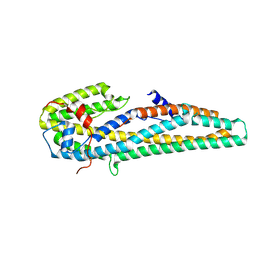 | | Crystal structure of trypsin activated Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Hey, T, Chikwana, V.M, Narva, K.E. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
5KUD
 
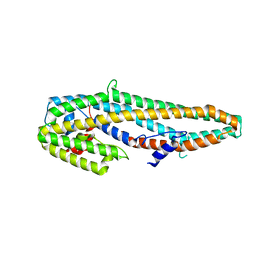 | | Crystal structure of full length Cry6Aa | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Kelker, M.S, Xu, X, Lee, M, Chan, M, Hung, S, Dementiev, K, Chikwana, V.M, Hey, T, Narva, K. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Bmc Biol., 14, 2016
|
|
5D65
 
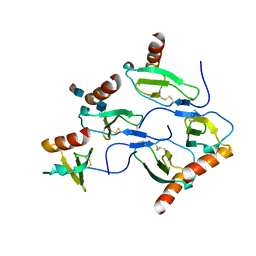 | | X-RAY STRUCTURE OF MACROPHAGE INFLAMMATORY PROTEIN-1 ALPHA (CCL3) WITH HEPARIN COMPLEX | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, C-C motif chemokine 3, CHLORIDE ION, ... | | Authors: | Liang, W.G, Hwang, D.Y, Zulueta, M.M, Hung, S.C, Tang, W. | | Deposit date: | 2015-08-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6LDK
 
 | |
