4TYE
 
 | | Structural analysis of the human Fibroblast Growth Factor Receptor 4 Kinase | | Descriptor: | Fibroblast growth factor receptor 4, PHOSPHATE ION | | Authors: | Lesca, E, Lammens, A, Huber, R, Augustin, M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of the human fibroblast growth factor receptor 4 kinase.
J.Mol.Biol., 426, 2014
|
|
4TYJ
 
 | | Structural analysis of the human Fibroblast Growth Factor Receptor 4 Kinase | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, Fibroblast growth factor receptor 4, ... | | Authors: | Lesca, E, Lammens, A, Huber, R, Augustin, M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural analysis of the human fibroblast growth factor receptor 4 kinase.
J.Mol.Biol., 426, 2014
|
|
2OQ5
 
 | | Crystal structure of DESC1, a new member of the type II transmembrane serine proteinases family | | Descriptor: | BENZAMIDINE, Transmembrane protease, serine 11E | | Authors: | Kyrieleis, O.J.P, Huber, R, Madison, E.L, Jacob, U. | | Deposit date: | 2007-01-31 | | Release date: | 2007-04-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of the catalytic domain of DESC1, a new member of the type II transmembrane serine proteinase family.
Febs J., 274, 2007
|
|
1JAE
 
 | | STRUCTURE OF TENEBRIO MOLITOR LARVAL ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Strobl, S, Maskos, K, Betz, M, Wiegand, G, Huber, R, Gomis-Rueth, F.X, Frank, G, Glockshuber, R. | | Deposit date: | 1997-09-30 | | Release date: | 1998-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of yellow meal worm alpha-amylase at 1.64 A resolution.
J.Mol.Biol., 278, 1998
|
|
2JDX
 
 | |
4F7S
 
 | | Crystal structure of human CDK8/CYCC in the DMG-in conformation | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-16 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F7J
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 3 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-(2-hydroxyethyl)urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-(2-hydroxyethyl)urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-16 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F6U
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 5 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[3-(morpholin-4-yl)propyl]urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[3-(morpholin-4-yl)propyl]urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F6W
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 1 (N-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-4-[2-({[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]carbamoyl}amino)ethyl]piperazine-1-carboxamide) | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F7L
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 2 (tert-butyl [3-({[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]carbamoyl}amino)propyl]carbamate) | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, tert-butyl [3-({[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]carbamoyl}amino)propyl]carbamate | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-16 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F70
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 4 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[2-(morpholin-4-yl)ethyl]urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[2-(morpholin-4-yl)ethyl]urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F7N
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 11 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-(5-hydroxypentyl)urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-(5-hydroxypentyl)urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-16 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4F6S
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 7 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4G6L
 
 | | Crystal structure of human CDK8/CYCC in the DMG-in conformation | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, FORMIC ACID | | Authors: | Schneider, E.V, Blaesse, M, Huber, R, Maskos, K. | | Deposit date: | 2012-07-19 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1EJA
 
 | | STRUCTURE OF PORCINE TRYPSIN COMPLEXED WITH BDELLASTASIN, AN ANTISTASIN-TYPE INHIBITOR | | Descriptor: | BDELLASTASIN, SODIUM ION, TRYPSIN | | Authors: | Rester, U, Moser, M, Huber, R, Bode, W. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | L-Isoaspartate 115 of porcine beta-trypsin promotes crystallization of its complex with bdellastasin.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3V3B
 
 | | Structure of the Stapled p53 Peptide Bound to Mdm2 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, SAH-p53-8 stapled-peptide | | Authors: | Baek, S, Kutchukian, P.S, Verdine, G.L, Huber, R, Holak, T.A, Ki Won, L, Popowicz, G.M. | | Deposit date: | 2011-12-13 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the stapled p53 peptide bound to Mdm2.
J.Am.Chem.Soc., 134, 2012
|
|
3V3V
 
 | | Structural and functional analysis of quercetagetin, a natural JNK1 inhibitor | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, C-Jun-amino-terminal kinase-interacting protein 1, CHLORIDE ION, ... | | Authors: | Baek, S, Kang, N.J, Popowicz, G.M, Arciniega, M, Jung, S.K, Byun, S, Song, N.R, Heo, Y.S, Kim, B.Y, Lee, H.J, Holak, T.A, Augustin, M, Bode, A.M, Huber, R, Dong, Z, Lee, K.W. | | Deposit date: | 2011-12-14 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of the Natural JNK1 Inhibitor Quercetagetin.
J.Mol.Biol., 425, 2013
|
|
1IXO
 
 | | Enzyme-analogue substrate complex of Pyridoxine 5'-Phosphate Synthase | | Descriptor: | Pyridoxine 5'-Phosphate synthase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
1IXQ
 
 | | Enzyme-Phosphate2 Complex of Pyridoxine 5'-Phosphate synthase | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-phosphate Synthase | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
1IXP
 
 | | Enzyme-phosphate Complex of Pyridoxine 5'-Phosphate synthase | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-Phosphate synthase | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
1IXN
 
 | | Enzyme-Substrate Complex of Pyridoxine 5'-Phosphate Synthase | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, Pyridoxine 5'-Phosphate Synthase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
1JC9
 
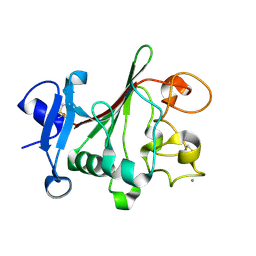 | | TACHYLECTIN 5A FROM TACHYPLEUS TRIDENTATUS (JAPANESE HORSESHOE CRAB) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, techylectin-5A | | Authors: | Kairies, N, Beisel, H.-G, Fuentes-Prior, P, Tsuda, R, Muta, T, Iwanaga, S, Bode, W, Huber, R, Kawabata, S. | | Deposit date: | 2001-06-08 | | Release date: | 2001-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The 2.0-A crystal structure of tachylectin 5A provides evidence for the common origin of the innate immunity and the blood coagulation systems.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1VLB
 
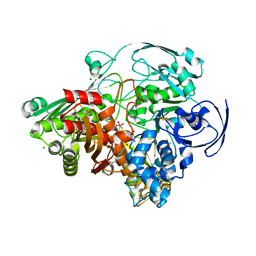 | | STRUCTURE REFINEMENT OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO GIGAS AT 1.28 A | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Rebelo, J.M, Dias, J.M, Huber, R, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2004-07-20 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure refinement of the aldehyde oxidoreductase from Desulfovibrio gigas (MOP) at 1.28 A
J.Biol.Inorg.Chem., 6, 2001
|
|
2QZW
 
 | |
6HP8
 
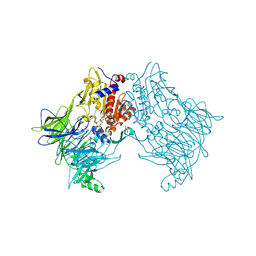 | | Crystal structure of DPP8 in complex with Val-BoroPro | | Descriptor: | Dipeptidyl peptidase 8, GLYCEROL, SODIUM ION, ... | | Authors: | Ross, B.H, Huber, R. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Improvement of Protein Crystal Diffraction Using Post-Crystallization Methods: Infrared Laser Radiation Controls Crystal Order
Thesis, 2019
|
|
