1UVS
 
 | | BOVINE THROMBIN--BM51.1011 COMPLEX | | Descriptor: | THROMBIN, [[CYCLOHEXANESULFONYL-GLYCYL]-3[PYRIDIN-4-YL-AMINOMETHYL]ALANYL]PIPERIDINE | | Authors: | Engh, R.A, Huber, R. | | Deposit date: | 1996-10-16 | | Release date: | 1997-11-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme flexibility, solvent and 'weak' interactions characterize thrombin-ligand interactions: implications for drug design.
Structure, 4, 1996
|
|
1UVU
 
 | | BOVINE THROMBIN--BM12.1700 COMPLEX | | Descriptor: | 3-(7-DIAMINOMETHYL-NAPHTHALEN-2-YL)-PROPIONIC ACID ETHYL ESTER, THROMBIN | | Authors: | Engh, R.A, Huber, R. | | Deposit date: | 1996-10-16 | | Release date: | 1997-11-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme flexibility, solvent and 'weak' interactions characterize thrombin-ligand interactions: implications for drug design.
Structure, 4, 1996
|
|
1Q62
 
 | | PKA double mutant model of PKB | | Descriptor: | cAMP-dependent protein kinase inhibitor, alpha form, cAMP-dependent protein kinase, ... | | Authors: | Gassel, M, Breitenlechner, C.B, Rueger, P, Jucknischke, U, Schneider, T, Huber, R, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2003-08-12 | | Release date: | 2003-09-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)
J.Mol.Biol., 329, 2003
|
|
1G8H
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE: THE TERNARY PRODUCT COMPLEX WITH APS AND PPI | | Descriptor: | ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, CADMIUM ION, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1G5Q
 
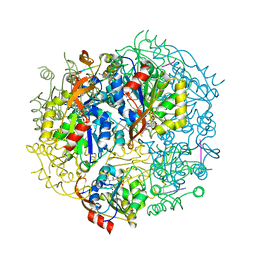 | | EPID H67N COMPLEXED WITH SUBSTRATE PEPTIDE DSYTC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, EPIDERMIN MODIFYING ENZYME EPID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbacher, S. | | Deposit date: | 2000-11-02 | | Release date: | 2001-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate.
EMBO J., 19, 2000
|
|
1ORV
 
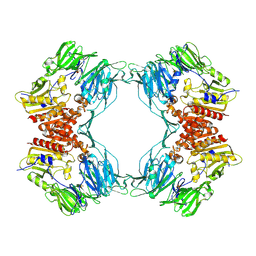 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1G63
 
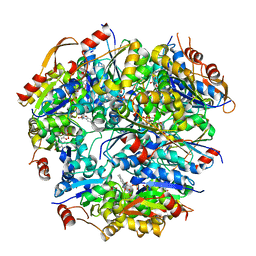 | | PEPTIDYL-CYSTEINE DECARBOXYLASE EPID | | Descriptor: | EPIDERMIN MODIFYING ENZYME EPID, FLAVIN MONONUCLEOTIDE | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbac, S. | | Deposit date: | 2000-11-03 | | Release date: | 2001-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate.
EMBO J., 19, 2000
|
|
1G71
 
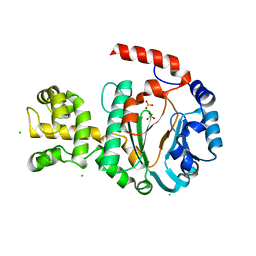 | | CRYSTAL STRUCTURE OF PYROCOCCUS FURIOSUS DNA PRIMASE | | Descriptor: | CHLORIDE ION, DNA PRIMASE, SULFATE ION, ... | | Authors: | Augustin, M.A, Huber, R, Kaiser, J.T. | | Deposit date: | 2000-11-08 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a DNA-dependent RNA polymerase (DNA primase).
Nat.Struct.Biol., 8, 2001
|
|
1PCA
 
 | | THREE DIMENSIONAL STRUCTURE OF PORCINE PANCREATIC PROCARBOXYPEPTIDASE A. A COMPARISON OF THE A AND B ZYMOGENS AND THEIR DETERMINANTS FOR INHIBITION AND ACTIVATION | | Descriptor: | CITRIC ACID, PROCARBOXYPEPTIDASE A PCPA, VALINE, ... | | Authors: | Guasch, A, Coll, M, Aviles, F.X, Huber, R. | | Deposit date: | 1991-10-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of porcine pancreatic procarboxypeptidase A. A comparison of the A and B zymogens and their determinants for inhibition and activation.
J.Mol.Biol., 224, 1992
|
|
1D02
 
 | | CRYSTAL STRUCTURE OF MUNI RESTRICTION ENDONUCLEASE IN COMPLEX WITH COGNATE DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*TP*TP*GP*GP*C)-3'), TYPE II RESTRICTION ENZYME MUNI | | Authors: | Deibert, M, Grazulis, S, Janulaitis, A, Siksnys, V, Huber, R. | | Deposit date: | 1999-09-08 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of MunI restriction endonuclease in complex with cognate DNA at 1.7 A resolution.
EMBO J., 18, 1999
|
|
1Q24
 
 | | PKA double mutant model of PKB in complex with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent Protein Kinase Inhibitor, ... | | Authors: | Gassel, M, Breitenlechner, C.B, Rueger, P, Jucknischke, U, Schneider, T, Huber, R, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2003-07-23 | | Release date: | 2003-08-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)
J.Mol.Biol., 329, 2003
|
|
1DD3
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L12 FROM THERMOTOGA MARITIMA | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12 | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
EMBO J., 19, 2000
|
|
1DGJ
 
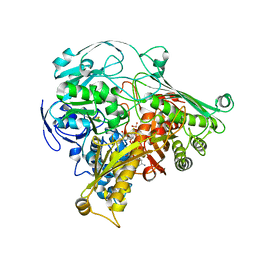 | | CRYSTAL STRUCTURE OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 | | Descriptor: | ALDEHYDE OXIDOREDUCTASE, FE2/S2 (INORGANIC) CLUSTER, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Rebelo, J.M, Macieira, S, Dias, J.M, Huber, R, Romao, M.J. | | Deposit date: | 1999-11-24 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Gene sequence and crystal structure of the aldehyde oxidoreductase from Desulfovibrio desulfuricans ATCC 27774.
J.Mol.Biol., 297, 2000
|
|
1DD4
 
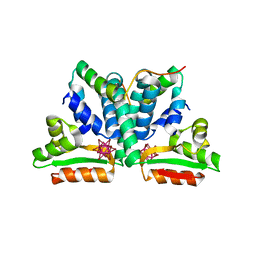 | | Crystal structure of ribosomal protein l12 from thermotoga maritim | | Descriptor: | 50S RIBOSOMAL PROTEIN L7/L12, HEXATANTALUM DODECABROMIDE | | Authors: | Wahl, M.C, Bourenkov, G.P, Bartunik, H.D, Huber, R. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flexibility, conformational diversity and two dimerization modes in complexes of ribosomal protein L12.
Embo J., 19, 2000
|
|
1ILS
 
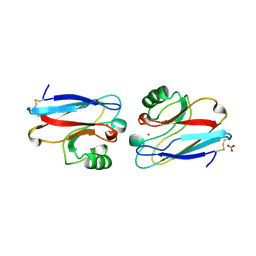 | | X-RAY CRYSTAL STRUCTURE THE TWO SITE-SPECIFIC MUTANTS ILE7SER AND PHE110SER OF AZURIN FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | AZURIN, COPPER (II) ION, NITRATE ION | | Authors: | Hammann, C, Nar, H, Huber, R, Messerschmidt, A. | | Deposit date: | 1995-10-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of the two site-specific mutants Ile7Ser and Phe110Ser of azurin from Pseudomonas aeruginosa.
J.Mol.Biol., 255, 1996
|
|
1JDX
 
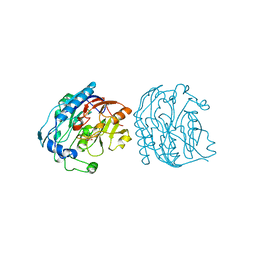 | |
1S4V
 
 | | The 2.0 A crystal structure of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm | | Descriptor: | DVA-LEU-LYS-0QE peptide, SULFATE ION, cysteine endopeptidase | | Authors: | Than, M.E, Helm, M, Simpson, D.J, Lottspeich, F, Huber, R, Gietl, C. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure and substrate specificity of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm.
J.Mol.Biol., 336, 2004
|
|
1U7Z
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, 4'-phosphopantothenoyl-CMP complex | | Descriptor: | Coenzyme A biosynthesis bifunctional protein coaBC, PHOSPHORIC ACID MONO-[3-(3-{[5-(4-AMINO-2-OXO-2H-PYRIMIDIN-1-YL)-3,4- DIHYDROXY-TETRAHYDRO-FURAN-2- YLMETHOXY]-HYDROXY-PHOSPHORYLOXY}-3-OXO-PROPYLCARBAMOYL)-3-HYDROXY-2,2- DIMETHYL-PROPYL] ESTER | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1U7U
 
 | | Phosphopantothenoylcysteine synthetase from E. coli | | Descriptor: | Coenzyme A biosynthesis bifunctional protein coaBC | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1U80
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, CMP complex | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC, PHOSPHATE ION | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1RU3
 
 | | Crystal Structure of the monomeric acetyl-CoA synthase from Carboxydothermus hydrogenoformans | | Descriptor: | Acetyl-CoA synthase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Svetlitchnyi, V, Dobbek, H, Meyer-Klaucke, W, Meins, T, Thiele, B, Rmer, P, Huber, R, Meyer, O. | | Deposit date: | 2003-12-11 | | Release date: | 2003-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A functional Ni-Ni-[4Fe-4S] cluster in the monomeric acetyl-CoA synthase from Carboxydothermus hydrogenoformans
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U7W
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, CTP-complex | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1RRX
 
 | | Crystallographic Evidence for Isomeric Chromophores in 3-Fluorotyrosyl-Green Fluorescent Protein | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Bae, J.H, Paramita Pal, P, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-12-09 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Evidence for Isomeric Chromophores in 3-Fluorotyrosyl-Green Fluorescent Protein.
Chembiochem, 5, 2004
|
|
1SM4
 
 | | Crystal Structure Analysis of the Ferredoxin-NADP+ Reductase from Paprika | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, chloroplast ferredoxin-NADP+ oxidoreductase | | Authors: | Dorowski, A, Hofmann, A, Steegborn, C, Boicu, M, Huber, R. | | Deposit date: | 2004-03-08 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of paprika ferredoxin-NADP+ reductase. Implications for the electron transfer pathway.
J.Biol.Chem., 276, 2001
|
|
1R6V
 
 | | Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin | | Descriptor: | CALCIUM ION, subtilisin-like serine protease | | Authors: | Kim, J.S, Kluskens, L.D, de Vos, W.M, Huber, R, van der Oost, J. | | Deposit date: | 2003-10-17 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin.
J.Mol.Biol., 335, 2004
|
|
