1KLO
 
 | |
1P8J
 
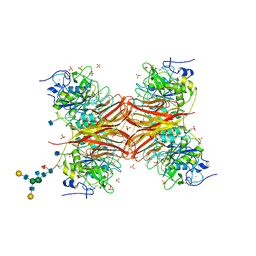 | | CRYSTAL STRUCTURE OF THE PROPROTEIN CONVERTASE FURIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DECANOYL-ARG-VAL-LYS-ARG-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Henrich, S, Cameron, A, Bourenkov, G.P, Kiefersauer, R, Huber, R, Lindberg, I, Bode, W, Than, M.E. | | Deposit date: | 2003-05-07 | | Release date: | 2003-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of the Proprotein Processing Proteinase Furin Explains its Stringent Specificity
Nat.Struct.Biol., 10, 2003
|
|
1CL2
 
 | |
1BLI
 
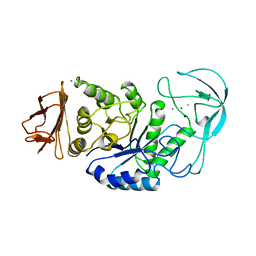 | | BACILLUS LICHENIFORMIS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 1998-01-07 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activation of Bacillus licheniformis alpha-amylase through a disorder-->order transition of the substrate-binding site mediated by a calcium-sodium-calcium metal triad.
Structure, 6, 1998
|
|
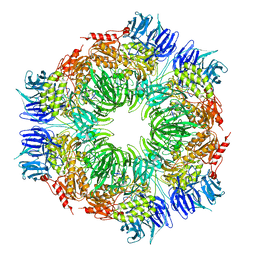
1G0U
 
 | | A GATED CHANNEL INTO THE PROTEASOME CORE PARTICLE | | Descriptor: | MAGNESIUM ION, PROTEASOME COMPONENT C1, PROTEASOME COMPONENT C11, ... | | Authors: | Groll, M, Bajorek, M, Kohler, A, Moroder, L, Rubin, D.M, Huber, R, Glickman, M.H, Finley, D. | | Deposit date: | 2000-10-09 | | Release date: | 2000-11-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A gated channel into the proteasome core particle.
Nat.Struct.Biol., 7, 2000
|
|
1N6E
 
 | | tricorn protease in complex with a tridecapeptide chloromethyl ketone derivative | | Descriptor: | DQTQKAAAELTFF, Tricorn protease | | Authors: | Kim, J.-S, Groll, M, Huber, R, Brandstetter, H. | | Deposit date: | 2002-11-10 | | Release date: | 2002-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Navigation Inside a Protease: Substrate Selection and Product Exit in the Tricorn Protease
from Thermoplasma acidophilum
J.Mol.Biol., 324, 2002
|
|
1J2Q
 
 | | 20S proteasome in complex with calpain-Inhibitor I from archaeoglobus fulgidus | | Descriptor: | 2-ACETYLAMINO-4-METHYL-PENTANOIC ACID [1-(1-FORMYL-PENTYLCARBAMOYL)-3-METHYL-BUTYL]-AMIDE, Proteasome alpha subunit, Proteasome beta subunit | | Authors: | Groll, M, Brandstetter, H, Bartunik, H, Bourenkow, G, Huber, R. | | Deposit date: | 2003-01-08 | | Release date: | 2003-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Investigations on the Maturation and Regulation of Archaebacterial Proteasomes
J.MOL.BIOL., 327, 2003
|
|
1N6D
 
 | | Tricorn protease in complex with tetrapeptide chloromethyl ketone derivative | | Descriptor: | DECANE, RVRK, Tricorn protease | | Authors: | Kim, J.-S, Groll, M, Huber, R, Brandstetter, H. | | Deposit date: | 2002-11-10 | | Release date: | 2002-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Navigation Inside a Protease: Substrate Selection and Product Exit in the Tricorn Protease
from Thermoplasma acidophilum
J.Mol.Biol., 324, 2002
|
|
1C7N
 
 | | CRYSTAL STRUCTURE OF CYSTALYSIN FROM TREPONEMA DENTICOLA CONTAINS A PYRIDOXAL 5'-PHOSPHATE COFACTOR | | Descriptor: | CYSTALYSIN, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Krupka, H.I, Huber, R, Holt, S.C, Clausen, T. | | Deposit date: | 2000-03-16 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5'-phosphate-dependent protein acting as a haemolytic enzyme.
EMBO J., 19, 2000
|
|
1C7O
 
 | | CRYSTAL STRUCTURE OF CYSTALYSIN FROM TREPONEMA DENTICOLA CONTAINS A PYRIDOXAL 5'-PHOSPHATE-L-AMINOETHOXYVINYLGLYCINE COMPLEX | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, CYSTALYSIN | | Authors: | Krupka, H.I, Huber, R, Holt, S.C, Clausen, T. | | Deposit date: | 2000-03-16 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5'-phosphate-dependent protein acting as a haemolytic enzyme.
EMBO J., 19, 2000
|
|
1GSS
 
 | | THREE-DIMENSIONAL STRUCTURE OF CLASS PI GLUTATHIONE S-TRANSFERASE FROM HUMAN PLACENTA IN COMPLEX WITH S-HEXYLGLUTATHIONE AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-hexyl-L-cysteinylglycine | | Authors: | Reinemer, P, Dirr, H.W, Ladenstein, R, Lobello, M, Federici, G, Huber, R, Parker, M.W. | | Deposit date: | 1992-05-28 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of class pi glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
1VPE
 
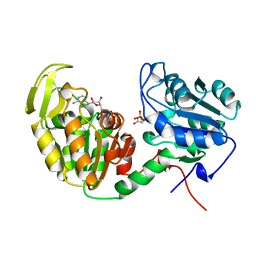 | | CRYSTALLOGRAPHIC ANALYSIS OF PHOSPHOGLYCERATE KINASE FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Auerbach, G, Huber, R, Graettinger, M, Zaiss, K, Schurig, H, Jaenicke, R, Jacob, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Closed structure of phosphoglycerate kinase from Thermotoga maritima reveals the catalytic mechanism and determinants of thermal stability.
Structure, 5, 1997
|
|
1DHP
 
 | | DIHYDRODIPICOLINATE SYNTHASE | | Descriptor: | DIHYDRODIPICOLINATE SYNTHASE, POTASSIUM ION | | Authors: | Mirwaldt, C, Korndoerfer, I, Huber, R. | | Deposit date: | 1995-02-09 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of dihydrodipicolinate synthase from Escherichia coli at 2.5 A resolution.
J.Mol.Biol., 246, 1995
|
|
1JEC
 
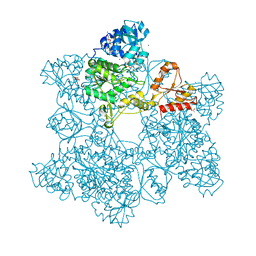 | |
1JED
 
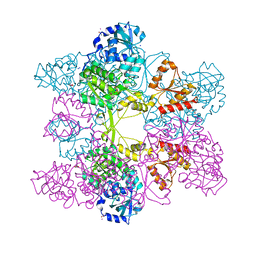 | | Crystal Structure of ATP Sulfurylase in complex with ADP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ullrich, T.C, Huber, R. | | Deposit date: | 2001-06-17 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The complex structures of ATP sulfurylase with thiosulfate, ADP and chlorate reveal new insights in inhibitory effects and the catalytic cycle.
J.Mol.Biol., 313, 2001
|
|
1JEE
 
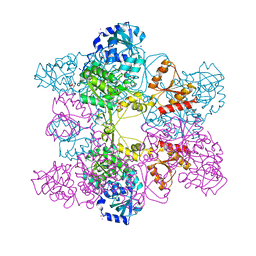 | | Crystal Structure of ATP Sulfurylase in complex with chlorate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Ullrich, T.C, Huber, R. | | Deposit date: | 2001-06-17 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The complex structures of ATP sulfurylase with thiosulfate, ADP and chlorate reveal new insights in inhibitory effects and the catalytic cycle.
J.Mol.Biol., 313, 2001
|
|
1RVV
 
 | | SYNTHASE/RIBOFLAVIN SYNTHASE COMPLEX OF BACILLUS SUBTILIS | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, PHOSPHATE ION, RIBOFLAVIN SYNTHASE | | Authors: | Ritsert, K, Huber, R, Turk, D, Ladenstein, R, Schmidt-Baese, K, Bacher, A. | | Deposit date: | 1995-10-25 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Studies on the lumazine synthase/riboflavin synthase complex of Bacillus subtilis: crystal structure analysis of reconstituted, icosahedral beta-subunit capsids with bound substrate analogue inhibitor at 2.4 A resolution.
J.Mol.Biol., 253, 1995
|
|
1N6F
 
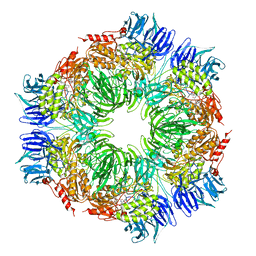 | | tricorn protease in complex with Z-Phe-diketo-Arg-Glu-Phe | | Descriptor: | 4-[2-(3-BENZYLOXYCARBONYLAMINO-4-CYCLOHEXYL-1-HYDROXY-2-OXO-BUTYLAMINO)-5-GUANIDINO-PENTANOYLAMINO]-4-(1-CARBOXY-2-CYCLOHEXYL-ETHYLCARBAMOYL)-BUTYRIC ACID, tricorn protease | | Authors: | Kim, J.-S, Groll, M, Huber, R, Brandstetter, H. | | Deposit date: | 2002-11-10 | | Release date: | 2002-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Navigation Inside a Protease: Substrate Selection and Product Exit in the Tricorn Protease
from Thermoplasma acidophilum
J.Mol.Biol., 324, 2002
|
|
1FBX
 
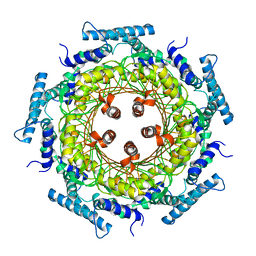 | | CRYSTAL STRUCTURE OF ZINC-CONTAINING E.COLI GTP CYCLOHYDROLASE I | | Descriptor: | CHLORIDE ION, GTP CYCLOHYDROLASE I, ZINC ION | | Authors: | Auerbach, G, Herrmann, A, Bracher, A, Bader, A, Gutlich, M, Fischer, M, Neukamm, M, Nar, H, Garrido-Franco, M, Richardson, J, Huber, R, Bacher, A. | | Deposit date: | 2000-07-17 | | Release date: | 2001-02-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Zinc plays a key role in human and bacterial GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1G8F
 
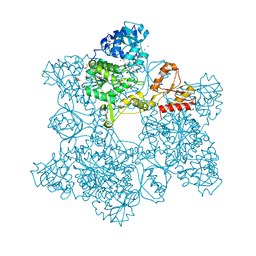 | | ATP SULFURYLASE FROM S. CEREVISIAE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, CADMIUM ION, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
1KYY
 
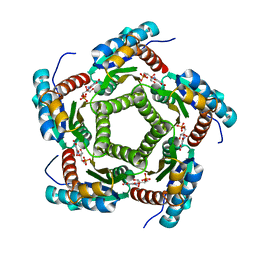 | | Lumazine Synthase from S.pombe bound to nitropyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1PPE
 
 | |
1G63
 
 | | PEPTIDYL-CYSTEINE DECARBOXYLASE EPID | | Descriptor: | EPIDERMIN MODIFYING ENZYME EPID, FLAVIN MONONUCLEOTIDE | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbac, S. | | Deposit date: | 2000-11-03 | | Release date: | 2001-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate.
EMBO J., 19, 2000
|
|
1UVS
 
 | | BOVINE THROMBIN--BM51.1011 COMPLEX | | Descriptor: | THROMBIN, [[CYCLOHEXANESULFONYL-GLYCYL]-3[PYRIDIN-4-YL-AMINOMETHYL]ALANYL]PIPERIDINE | | Authors: | Engh, R.A, Huber, R. | | Deposit date: | 1996-10-16 | | Release date: | 1997-11-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme flexibility, solvent and 'weak' interactions characterize thrombin-ligand interactions: implications for drug design.
Structure, 4, 1996
|
|
1G8G
 
 | | ATP SULFURYLASE FROM S. CEREVISIAE: THE BINARY PRODUCT COMPLEX WITH APS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Ullrich, T.C, Blaesse, M, Huber, R. | | Deposit date: | 2000-11-17 | | Release date: | 2001-05-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation.
EMBO J., 20, 2001
|
|
