7ZBR
 
 | | Crystal Structure of full-length Bartonella Effector protein 1 | | Descriptor: | Bartonella effector protein (Bep) substrate of VirB T4SS | | Authors: | Huber, M. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Full-length structure of a FIC effector protein from Bartonella reveals a novel structural domain
To be published
|
|
2L3Z
 
 | | Proton-Detected 4D DREAM Solid-State NMR Structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Huber, M, Hiller, S, Schanda, P, Ernst, M, Bockmann, A, Verel, R, Meier, B.H. | | Deposit date: | 2010-09-27 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | A Proton-Detected 4D Solid-State NMR Experiment for Protein Structure Determination.
Chemphyschem, 12, 2011
|
|
5OQK
 
 | | Solution NMR structure of truncated, human Hv1/VSOP (Voltage-gated proton channel) | | Descriptor: | Voltage-gated hydrogen channel 1 | | Authors: | Bayrhuber, M, Maslennikov, I, Kwiatowski, W, Sobol, A, Wierschem, C, Eichmann, C, Riek, R. | | Deposit date: | 2017-08-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure and Functional Behavior of the Human Proton Channel.
Biochemistry, 58, 2019
|
|
2JK4
 
 | | Structure of the human voltage-dependent anion channel | | Descriptor: | VOLTAGE-DEPENDENT ANION-SELECTIVE CHANNEL PROTEIN 1 | | Authors: | Bayrhuber, M, Meins, T, Habeck, M, Becker, S, Giller, K, Villinger, S, Vonrhein, C, Griesinger, C, Zweckstetter, M, Zeth, K. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of the Human Voltage-Dependent Anion Channel.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2CA7
 
 | | Conkunitzin-S1 Is The First Member Of A New Kunitz-Type Neurotoxin Family- Structural and Functional Characterization | | Descriptor: | CONKUNITZIN-S1 | | Authors: | Bayrhuber, M, Vijayan, V, Ferber, M, Graf, R, Korukottu, J, Imperial, J, Garrett, J.E, Olivera, B.M, Terlau, H, Zweckstetter, M, Becker, S. | | Deposit date: | 2005-12-19 | | Release date: | 2006-01-05 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Conkunitzin-S1 is the first member of a new Kunitz-type neurotoxin family. Structural and functional characterization.
J. Biol. Chem., 280, 2005
|
|
2LIE
 
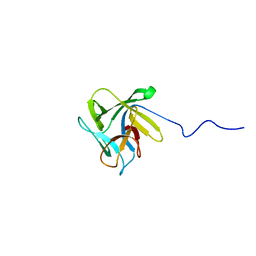 | | NMR structure of the lectin CCL2 | | Descriptor: | CCL2 lectin | | Authors: | Schubert, M, Walti, M.A, Egloff, P, Bleuler-Martinez, S, Aebi, M, Allain, F.F.-H, Kunzler, M. | | Deposit date: | 2011-08-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
2LIQ
 
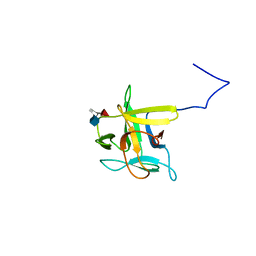 | | Solution structure of CCL2 in complex with glycan | | Descriptor: | CCL2 lectin, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Schubert, M, Bleuler-Martinez, S, Walti, M.A, Egloff, P, Aebi, M, Kuenzler, M, Allain, F.H.-T. | | Deposit date: | 2011-08-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
2MK1
 
 | |
1SN8
 
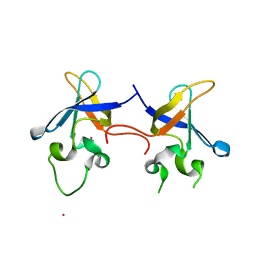 | | Crystal structure of the S1 domain of RNase E from E. coli (Pb derivative) | | Descriptor: | LEAD (II) ION, Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1SMX
 
 | | Crystal structure of the S1 domain of RNase E from E. coli (native) | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1SLJ
 
 | | Solution structure of the S1 domain of RNase E from E. coli | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
2LOR
 
 | | Backbone structure of human membrane protein TMEM141 | | Descriptor: | Transmembrane protein 141 | | Authors: | Bayrhuber, M, Klammt, C, Maslennikov, I, Riek, R, Choe, S. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Facile backbone structure determination of human membrane proteins by NMR spectroscopy.
Nat.Methods, 9, 2012
|
|
2MVX
 
 | | Atomic-resolution 3D structure of amyloid-beta fibrils: the Osaka mutation | | Descriptor: | Amyloid beta A4 protein | | Authors: | Schuetz, A.K, Vagt, T, Huber, M, Ovchinnikova, O.Y, Cadalbert, R, Wall, J, Guentert, P, Bockmann, A, Glockshuber, R, Meier, B.H. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Three-Dimensional Structure of Amyloid beta Fibrils Bearing the Osaka Mutation.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4USO
 
 | | X-ray structure of the CCL2 lectin in complex with sialyl lewis X | | Descriptor: | CCL2 LECTIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
3OAU
 
 | | Antibody 2G12 Recognizes Di-Mannose Equivalently in Domain- and Non-Domain-Exchanged Forms, but only binds the HIV-1 Glycan Shield if Domain-Exchanged | | Descriptor: | Fab 2G12, heavy chain, light chain, ... | | Authors: | Doores, K.J, Fulton, Z, Huber, M, Wilson, I.A, Burton, D.R. | | Deposit date: | 2010-08-05 | | Release date: | 2011-01-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged.
J.Virol., 84, 2010
|
|
2JBO
 
 | | Protein kinase MK2 in complex with an inhibitor (crystal form-1, soaking) | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP KINASE-ACTIVATED PROTEIN KINASE 2, PHOSPHATE ION | | Authors: | Hillig, R.C, Eberspaecher, U, Monteclaro, F, Huber, M, Nguyen, D, Mengel, A, Muller-Tiemann, B, Egner, U. | | Deposit date: | 2006-12-09 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for a High Affinity Inhibitor Bound to Protein Kinase Mk2.
J.Mol.Biol., 369, 2007
|
|
2JBP
 
 | | Protein kinase MK2 in complex with an inhibitor (crystal form-2, co- crystallization) | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP KINASE-ACTIVATED PROTEIN KINASE 2 | | Authors: | Hillig, R.C, Eberspaecher, U, Monteclaro, F, Huber, M, Nguyen, D, Mengel, A, Muller-Tiemann, B, Egner, U. | | Deposit date: | 2006-12-09 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis for a high affinity inhibitor bound to protein kinase MK2.
J. Mol. Biol., 369, 2007
|
|
1NPJ
 
 | | Crystal structure of H145A mutant of nitrite reductase from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, Boulanger, M.J, Molon, A, Fittipaldi, M, Huber, M, Murphy, M.E, Verbeet, M.P, Canters, G.W. | | Deposit date: | 2003-01-18 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reconstitution of the type-1 active site of the H145G/A variants of nitrite reductase by ligand insertion
Biochemistry, 42, 2003
|
|
1NPN
 
 | | Crystal structure of a copper reconstituted H145A mutant of nitrite reductase from Alcaligenes faecalis | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, Boulanger, M.J, Molon, A, Fittipaldi, M, Huber, M, Murphy, M.E, Verbeet, M.P, Canters, G.W. | | Deposit date: | 2003-01-18 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reconstitution of the type-1 active site of the 145G/A variants of Nitrite Reductase by ligand insertion
Biochemistry, 42, 2003
|
|
4USP
 
 | | X-ray structure of the dimeric CCL2 lectin in native form | | Descriptor: | CCL2 LECTIN, CHLORIDE ION, PHOSPHATE ION | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
8A0C
 
 | | Capsular polysaccharide synthesis multienzyme in complex with CMP | | Descriptor: | Bcs3, CYTIDINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Cifuente, J.O, Schulze, J, Bethe, A, Di Domenico, V, Litschko, C, Budde, I, Eidenberger, L, Thiesler, H, Ramon-Roth, I, Berger, M, Claus, H, DAngelo, C, Marina, A, Gerardy-Schahn, R, Schubert, M, Guerin, M.E, Fiebig, T. | | Deposit date: | 2022-05-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A multi-enzyme machine polymerizes the Haemophilus influenzae type b capsule.
Nat.Chem.Biol., 19, 2023
|
|
8A0M
 
 | | Capsular polysaccharide synthesis multienzyme in complex with capsular polymer fragment | | Descriptor: | Bcs3, MAGNESIUM ION, beta-D-ribosyl-(1->1)-D-ribitol-5-phosphate | | Authors: | Cifuente, J.O, Schulze, J, Bethe, A, Di Domenico, V, Litschko, C, Budde, I, Eidenberger, L, Thiesler, H, Ramon-Roth, I, Berger, M, Claus, H, DAngelo, C, Marina, A, Gerardy-Schahn, R, Schubert, M, Guerin, M.E, Fiebig, T. | | Deposit date: | 2022-05-29 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A multi-enzyme machine polymerizes the Haemophilus influenzae type b capsule.
Nat.Chem.Biol., 19, 2023
|
|
6EU9
 
 | | Crystal structure of Platynereis dumerilii RAR ligand-binding domain in complex with all-trans retinoic acid | | Descriptor: | RETINOIC ACID, Retinoic acid receptor | | Authors: | Handberg-Thorsager, M, Gutierrez-Mazariegos, J, Arold, S.T, Nadendla, E.K, Bertucci, P.Y, Germain, P, Tomancak, P, Pierzchalski, K, Jones, J.W, Albalat, R, Kane, M.A, Bourguet, W, Laudet, V, Arendt, D, Schubert, M. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The ancestral retinoic acid receptor was a low-affinity sensor triggering neuronal differentiation.
Sci Adv, 4, 2018
|
|
6EZ0
 
 | | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding | | Descriptor: | RNA (27-MER) | | Authors: | Erat, M.C, Besic, E, Oberhuber, M, Johannsen, S, Sigel, R.K.O. | | Deposit date: | 2017-11-13 | | Release date: | 2018-01-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Specific phosphorothioate substitution within domain 6 of a group II intron ribozyme leads to changes in local structure and metal ion binding.
J. Biol. Inorg. Chem., 23, 2018
|
|
2J6D
 
 | | CONKUNITZIN-S2 - CONE SNAIL NEUROTOXIN - DENOVO STRUCTURE | | Descriptor: | CONKUNITZIN-S2 | | Authors: | Korukottu, J, Bayrhuber, M, Montaville, P, Vijayan, V, Jung, Y.-S, Becker, S, Zweckstetter, M. | | Deposit date: | 2006-09-27 | | Release date: | 2007-01-16 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Fast High-Resolution Protein Structure Determination by Using Unassigned NMR Data.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
