5NRX
 
 | | Cys-Gly dipeptidase GliJ in complex with Fe2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, FE (III) ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NS2
 
 | | Cys-Gly dipeptidase GliJ in complex with Co2+ | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Dipeptidase gliJ, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NS1
 
 | | Cys-Gly dipeptidase GliJ in complex with Ni2+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, NICKEL (II) ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
5NRY
 
 | | Cys-Gly dipeptidase GliJ in complex with Fe3+ | | Descriptor: | CHLORIDE ION, Dipeptidase gliJ, FE (III) ION | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Gliotoxin Biosynthesis: Structure, Mechanism, and Metal Promiscuity of Carboxypeptidase GliJ.
ACS Chem. Biol., 12, 2017
|
|
4QLT
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 2 (PR924) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(3-methyl-1H-inden-2-yl)carbonyl]-D-alanyl-N-[(2S,4R)-5-hydroxy-4-methyl-3-oxo-1-phenylpentan-2-yl]-L-tryptophanamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
4QLQ
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-(morpholin-4-ylacetyl)-L-alanyl-N-[(2S,4R)-1-cyclohexyl-5-hydroxy-4-methyl-3-oxopentan-2-yl]-O-methyl-L-tyrosinamide, ... | | Authors: | De Bruin, G, Huber, E, Xin, B, Van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, Van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
4QLS
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 11 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-(morpholin-4-ylacetyl)-D-alanyl-N-[(2S,4R)-1-cyclohexyl-5-hydroxy-4-methyl-3-oxopentan-2-yl]-O-methyl-L-tyrosinamide, ... | | Authors: | De Bruin, G, Huber, E, Xin, B, Van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, Van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
4QLU
 
 | | yCP in complex with tripeptidic epoxyketone inhibitor 9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(3-methyl-1H-inden-2-yl)carbonyl]-D-alanyl-N-[(2S,4R)-1-cyclohexyl-5-hydroxy-4-methyl-3-oxopentan-2-yl]-L-tryptophanamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
4NTD
 
 | | Crystal structure of HlmI | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, ... | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NTE
 
 | | Crystal structure of DepH | | Descriptor: | CHLORIDE ION, DepH, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NTC
 
 | | Crystal structure of GliT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GliT | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6SUP
 
 | | Crystal Structure of TcdB2-TccC3-Cdc42 | | Descriptor: | MAGNESIUM ION, TcdB2,TccC3,Cell division control protein 42 homolog | | Authors: | Roderer, D, Schubert, E, Sitsel, O, Raunser, S. | | Deposit date: | 2019-09-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Towards the application of Tc toxins as a universal protein translocation system.
Nat Commun, 10, 2019
|
|
6SUQ
 
 | | Crystal Structure of TcdB2-TccC3-TEV | | Descriptor: | TcdB2,TccC3,Genome polyprotein | | Authors: | Roderer, D, Schubert, E, Sitsel, O, Raunser, S. | | Deposit date: | 2019-09-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Towards the application of Tc toxins as a universal protein translocation system.
Nat Commun, 10, 2019
|
|
6H6F
 
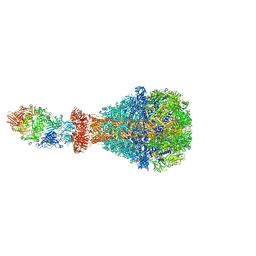 | | PTC3 holotoxin complex from Photorhabdus luminiscens - Mutant TcC-D651A | | Descriptor: | TcdA1, TcdB2,TccC3,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
6H6E
 
 | | PTC3 holotoxin complex from Photorhabdus luminecens in prepore state (TcdA1, TcdB2, TccC3) | | Descriptor: | TcdA1, TcdB2,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
6H6G
 
 | | Crystal Structure of TcdB2-TccC3 without hypervariable C-terminal region | | Descriptor: | TcdB2,TccC3 | | Authors: | Gatsogiannis, C, Merino, F, Roderer, D, Balchin, D, Schubert, E, Kuhlee, A, Hayer-Hartl, M, Raunser, S. | | Deposit date: | 2018-07-27 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Tc toxin activation requires unfolding and refolding of a beta-propeller.
Nature, 563, 2018
|
|
1ZFJ
 
 | | INOSINE MONOPHOSPHATE DEHYDROGENASE (IMPDH; EC 1.1.1.205) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | INOSINE MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID | | Authors: | Zhang, R, Evans, G, Rotella, F.J, Westbrook, E.M, Beno, D, Huberman, E, Joachimiak, A, Collart, F.R. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase.
Biochemistry, 38, 1999
|
|
