1EAI
 
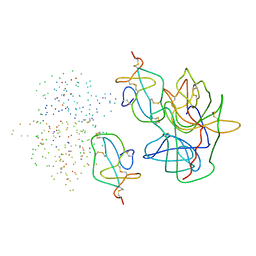 | | COMPLEX OF ASCARIS CHYMOTRPSIN/ELASTASE INHIBITOR WITH PORCINE ELASTASE | | Descriptor: | PROTEIN (CHYMOTRYPSIN/ELASTASE ISOINHIBITOR 1), PROTEIN (ELASTASE) | | Authors: | Huang, K, Strynadka, N.C.J, Bernard, V.D, Peanasky, R.J, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 1999-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of the complex of Ascaris chymotrypsin/elastase inhibitor with porcine elastase.
Structure, 2, 1994
|
|
2SGP
 
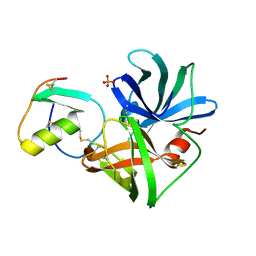 | | PRO 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | OVOMUCOID INHIBITOR, PHOSPHATE ION, PROTEINASE B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
2SGQ
 
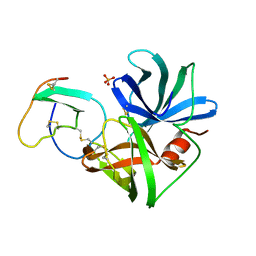 | | GLN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
2SGF
 
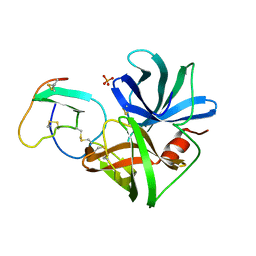 | | PHE 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
2SGE
 
 | | GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | Descriptor: | Ovomucoid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water molecules participate in proteinase-inhibitor interactions: crystal structures of Leu18, Ala18, and Gly18 variants of turkey ovomucoid inhibitor third domain complexed with Streptomyces griseus proteinase B.
Protein Sci., 4, 1995
|
|
7D23
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with one K ion bound to the active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutaminyl-peptide cyclotransferase, POTASSIUM ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1P
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with three Cd ions bound to the active site | | Descriptor: | BICARBONATE ION, CADMIUM ION, Glutaminyl-peptide cyclotransferase | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1B
 
 | | Crystal structure of Fimbriiglobus ruber glutaminyl cyclase | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D18
 
 | | Crystal structure of Acidobacteriales bacterium glutaminyl cyclase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Peptidase M28, ... | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1D
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutaminyl cyclase bound to 1-benzylimidazole | | Descriptor: | 1-BENZYL-1H-IMIDAZOLE, Glutamine cyclotransferase, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2B
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Ni ion bound to the active site | | Descriptor: | Glutaminyl-peptide cyclotransferase, NICKEL (II) ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2I
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Fe ion bound to the active site | | Descriptor: | FE (III) ION, Glutaminyl-peptide cyclotransferase, SULFATE ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1Y
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with two Co ions bound to the active site | | Descriptor: | COBALT (II) ION, Glutaminyl-peptide cyclotransferase | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D21
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with two Zn ions bound to the active site | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D17
 
 | | Crystal structure of Macrostomum lignano glutaminyl cyclase | | Descriptor: | Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1E
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutaminyl cyclase bound to N-acetylhistamine | | Descriptor: | Leucine aminopeptidase, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1N
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with three Cu ions bound to the active site | | Descriptor: | BICARBONATE ION, COPPER (II) ION, Glutaminyl-peptide cyclotransferase | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1H
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with D238A mutation | | Descriptor: | Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2J
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Cd ion bound to the active site | | Descriptor: | BICARBONATE ION, CADMIUM ION, Glutaminyl-peptide cyclotransferase | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2D
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Mn ion bound to the active site | | Descriptor: | Glutaminyl-peptide cyclotransferase, MANGANESE (II) ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
2SGD
 
 | | ASP 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | Descriptor: | Ovomucoid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
1PYM
 
 | |
2DIK
 
 | | R337A MUTANT OF PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | PROTEIN (PYRUVATE PHOSPHATE DIKINASE), SULFATE ION | | Authors: | Huang, K, Herzberg, O. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Location of the phosphate binding site within Clostridium symbiosum pyruvate phosphate dikinase.
Biochemistry, 37, 1998
|
|
3PB4
 
 | | Crystal structure of the catalytic domain of human Golgi-resident glutaminyl cyclase at pH 6.0 | | Descriptor: | Glutaminyl-peptide cyclotransferase-like protein, ZINC ION | | Authors: | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
3PBB
 
 | | Crystal structure of human secretory glutaminyl cyclase in complex with PBD150 | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Huang, K.F, Liaw, S.S, Huang, W.L, Chia, C.Y, Lo, Y.C, Chen, Y.L, Wang, A.H.J. | | Deposit date: | 2010-10-20 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of human Golgi-resident glutaminyl cyclase and its complexes with inhibitors reveal a large loop movement upon inhibitor binding
J.Biol.Chem., 286, 2011
|
|
