3LM1
 
 | | Crystal Structure Analysis of Maclura pomifera agglutinin complex with p-nitrophenyl-GalNAc | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Agglutinin alpha chain, Agglutinin beta-2 chain | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
3LLZ
 
 | | Crystal Structure Analysis of Maclura pomifera agglutinin complex with Gal-beta-1,3-GalNAc | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-2 chain, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
3LLY
 
 | | Crystal Structure Analysis of Maclura pomifera agglutinin | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-2 chain | | Authors: | Huang, J, Xu, Z, Wang, D, Ogato, C, Hirama, T, Palczewski, K, Hazen, S.L, Lee, X, Young, N.M. | | Deposit date: | 2010-01-29 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses.
Glycobiology, 20, 2010
|
|
3U85
 
 | |
3U86
 
 | |
5HIN
 
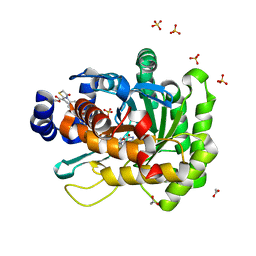 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 18L compound | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Huang, J, Wu, D, Lu, Q, Yao, X. | | Deposit date: | 2016-01-12 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 18L at 1.60 A resolution
To Be Published
|
|
5HQE
 
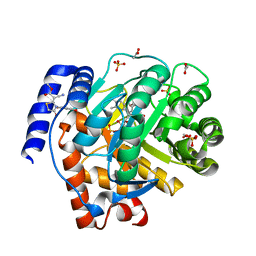 | |
5H73
 
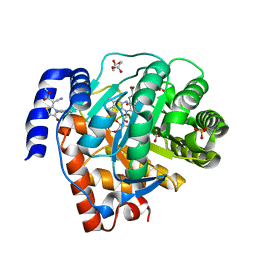 | | Crystal structure of human DHODH with 18F | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Huang, J, Wu, D. | | Deposit date: | 2016-11-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of human DHODH with 18F at 1.58 Angstroms resolution
To Be Published
|
|
5GHE
 
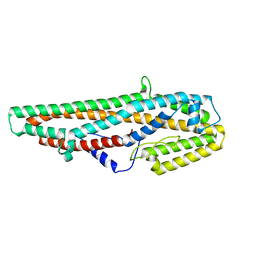 | | Crystal Structure of Bacillus thuringiensis Cry6Aa2 Protoxin | | Descriptor: | Pesticidal crystal protein Cry6Aa | | Authors: | Huang, J, Guan, Z, Zou, T, Sun, M, Yin, P. | | Deposit date: | 2016-06-19 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal structure of Cry6Aa: A novel nematicidal ClyA-type alpha-pore-forming toxin from Bacillus thuringiensis
Biochem.Biophys.Res.Commun., 478, 2016
|
|
7VOH
 
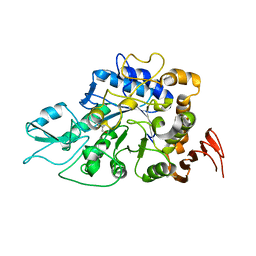 | | The a-glucosidase QsGH13 from Qipengyuania seohaensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, alpha-glucosidase QsGH13 | | Authors: | Huang, J, Zhai, X.Y. | | Deposit date: | 2021-10-13 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Structure and Function Insight of the alpha-Glucosidase QsGH13 From Qipengyuania seohaensis sp. SW-135.
Front Microbiol, 13, 2022
|
|
6KIX
 
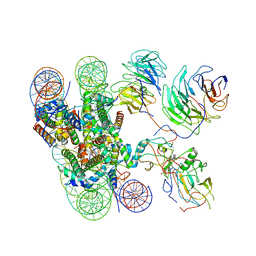 | | Cryo-EM structure of human MLL1-NCP complex, binding mode1 | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIV
 
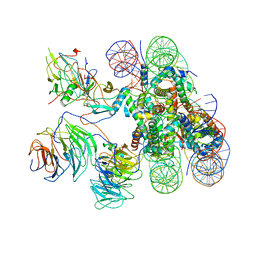 | | Cryo-EM structure of human MLL1-ubNCP complex (4.0 angstrom) | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIZ
 
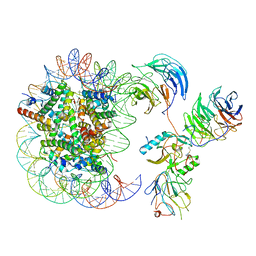 | | Cryo-EM structure of human MLL1-NCP complex, binding mode2 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIU
 
 | | Cryo-EM structure of human MLL1-ubNCP complex (3.2 angstrom) | | Descriptor: | DNA (145-MER), GLUTAMINE, Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
6KIW
 
 | | Cryo-EM structure of human MLL3-ubNCP complex (4.0 angstrom) | | Descriptor: | DNA (144-MER), DNA (145-MER), Histone H2A, ... | | Authors: | Huang, J, Xue, H, Yao, T. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of nucleosome recognition and modification by MLL methyltransferases.
Nature, 573, 2019
|
|
7JMK
 
 | |
7JFP
 
 | | GTP-specific succinyl-CoA synthetase complexed with Mg-GDP, phosphohistidine loop pointing towards nucleotide binding site | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Second distinct conformation of the phosphohistidine loop in succinyl-CoA synthetase
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7JKR
 
 | | GTP-specific succinyl-CoA synthetase complexed with Mg-GMPPNP, phosphohistidine loop pointing towards nucleotide binding site | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2020-07-28 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Second distinct conformation of the phosphohistidine loop in succinyl-CoA synthetase
Acta Crystallogr.,Sect.D, 77, 2021
|
|
7JJ0
 
 | | GTP-specific succinyl-CoA synthetase complexed with Mg-GMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, ... | | Authors: | Huang, J, Fraser, M.E. | | Deposit date: | 2020-07-24 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Second distinct conformation of the phosphohistidine loop in succinyl-CoA synthetase
Acta Crystallogr.,Sect.D, 77, 2021
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
8JMM
 
 | | Structure of XBB spike protein (S) dimer-trimer in complex with bispecific antibody G7-Fc at 3.75 Angstroms resolution. | | Descriptor: | 7F3 Fv, GW01 Fv, Spike glycoprotein | | Authors: | Hao, A, Mao, Q, Chen, Z, Huang, J, Sun, L. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of XBB spike protein (S) dimer-trimer in complex with bispecific antibody G7-Fc at 3.75 Angstroms resolution.
To Be Published
|
|
5WXH
 
 | | Crystal structure of TAF3 PHD finger bound to H3K4me3 | | Descriptor: | Histone H3K4me3, Transcription initiation factor TFIID subunit 3, ZINC ION | | Authors: | Zhao, S, Huang, J, Li, H. | | Deposit date: | 2017-01-07 | | Release date: | 2017-08-16 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.297 Å) | | Cite: | Kinetic and high-throughput profiling of epigenetic interactions by 3D-carbene chip-based surface plasmon resonance imaging technology
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3KVO
 
 | | Crystal structure of the catalytic domain of human Hydroxysteroid dehydrogenase like 2 (HSDL2) | | Descriptor: | Hydroxysteroid dehydrogenase-like protein 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ugochukwu, E, Bhatia, C, Huang, J, Pilka, E, Muniz, J.R.C, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Verdin, E.M, Oppermann, U, Kavanagh, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the catalytic domain of human Hydroxysteroid dehydrogenase like 2 (HSDL2)
To be Published
|
|
3U4V
 
 | |
3U4Z
 
 | |
