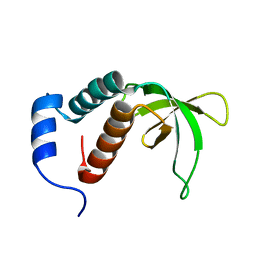
2P0P
 
 | |
6KUZ
 
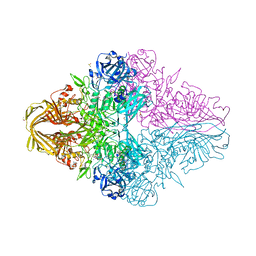 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSL01 | | 分子名称: | 3-(1,3-benzothiazol-2-yl)-2-[[4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]methoxy]-5-methyl-benzaldehyde, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | 著者 | Chen, X, Hu, Y.L, Li, X.K, Guo, Y, Li, J. | | 登録日 | 2019-09-03 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | First-generation species-selective chemical probes for fluorescence imaging of human senescence-associated beta-galactosidase.
Chem Sci, 11, 2020
|
|
8DGB
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Q192T Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-23 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DCZ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) M165Y Mutant in Complex with Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
1LI4
 
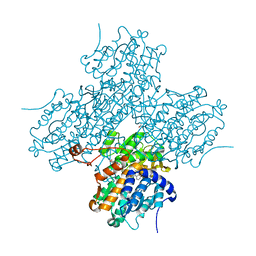 | | Human S-adenosylhomocysteine hydrolase complexed with neplanocin | | 分子名称: | 3-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-CYCLOPENTANE-1,2-DIOL, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Yang, X, Hu, Y, Yin, D.H, Turner, M.A, Wang, M, Borchardt, R.T, Howell, P.L, Kuczera, K, Schowen, R.L. | | 登録日 | 2002-04-17 | | 公開日 | 2003-05-20 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Catalytic strategy of S-adenosyl-L-homocysteine hydrolase: Transition-state
stabilization and the avoidance of abortive reactions
Biochemistry, 42, 2003
|
|
8DFN
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H164N Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
4G3H
 
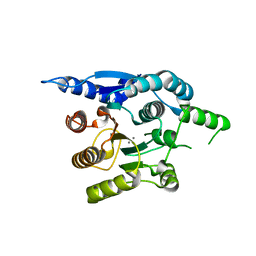 | | Crystal structure of helicobacter pylori arginase | | 分子名称: | Arginase (RocF), MANGANESE (II) ION | | 著者 | Zhang, J, Zhang, X, Li, D, Hu, Y, Zou, Q, Wang, D. | | 登録日 | 2012-07-13 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and function studies on Helicobacter pylori arginase
To be Published
|
|
8DD1
 
 | | SARS-CoV-2 Main Protease (Mpro) H164N Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFE
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-22 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD9
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant in Complex with Inhibitor GC376 | | 分子名称: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-17 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
7D11
 
 | |
6UG0
 
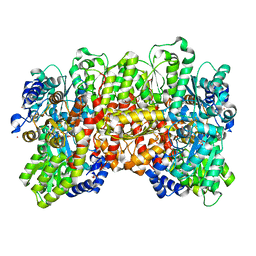 | | N2-bound Nitrogenase MoFe-protein from Azotobacter vinelandii | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Kang, W, Hu, Y, Ribbe, M.W. | | 登録日 | 2019-09-25 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
4D8M
 
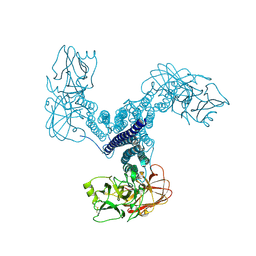 | | Crystal structure of Bacillus thuringiensis Cry5B nematocidal toxin | | 分子名称: | Pesticidal crystal protein cry5Ba | | 著者 | Fan, H, Hu, Y, Aroian, R.V, Ghosh, P, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2012-01-10 | | 公開日 | 2012-12-19 | | 最終更新日 | 2013-02-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and Glycolipid Binding Properties of the Nematicidal Protein Cry5B.
Biochemistry, 51, 2012
|
|
8EHM
 
 | |
8EHJ
 
 | |
5WYO
 
 | | Solution structure of E.coli HdeA | | 分子名称: | Acid stress chaperone HdeA | | 著者 | Yang, C, Hu, Y, Jin, C. | | 登録日 | 2017-01-14 | | 公開日 | 2017-11-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Characterizations of the Interactions between Escherichia coli Periplasmic Chaperone HdeA and Its Native Substrates during Acid Stress
Biochemistry, 56, 2017
|
|
6VXT
 
 | | Activated Nitrogenase MoFe-protein from Azotobacter vinelandii | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Kang, W, Hu, Y, Ribbe, M.W. | | 登録日 | 2020-02-24 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
3LKX
 
 | | Human nac dimerization domain | | 分子名称: | Nascent polypeptide-associated complex subunit alpha, Transcription factor BTF3 | | 著者 | Liu, Y, Hu, Y, Li, X, Niu, L, Teng, M. | | 登録日 | 2010-01-28 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit
Biochemistry, 49, 2010
|
|
1XS3
 
 | | Solution Structure Analysis of the XC975 protein | | 分子名称: | hypothetical protein XC975 | | 著者 | Chin, K.-H, Lin, F.-Y, Hu, Y.-C, Sze, K.-H, Lyu, P.-C, Chou, S.-H. | | 登録日 | 2004-10-18 | | 公開日 | 2005-03-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Letter to the Editor: NMR structure note - Solution structure of a bacterial BolA-like protein XC975 from a plant pathogen Xanthomonas campestris pv. campestris
J.Biomol.Nmr, 31, 2005
|
|
2MOK
 
 | | holo_FldA | | 分子名称: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | 著者 | Jin, C, Hu, Y, Ye, Q. | | 登録日 | 2014-04-27 | | 公開日 | 2015-05-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR study of YqcA from Escherichia coli
To be Published
|
|
8IEX
 
 | | Solution structure of AtWRKY11-DBD | | 分子名称: | Probable WRKY transcription factor 11, ZINC ION | | 著者 | Dong, X, Hu, Y.F. | | 登録日 | 2023-02-16 | | 公開日 | 2024-02-21 | | 最終更新日 | 2024-09-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the DNA binding domain of Arabidopsis transcription factor WRKY11.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
8D4L
 
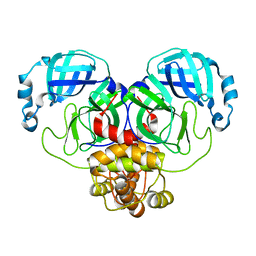 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144A Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8GQ6
 
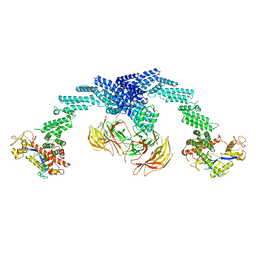 | | Cryo-EM Structure of the KBTBD2-CUL3-Rbx1 dimeric complex | | 分子名称: | Cullin-3, E3 ubiquitin-protein ligase RBX1, Kelch repeat and BTB domain-containing protein 2, ... | | 著者 | Sun, L, Chen, Z, Hu, Y, Mao, Q. | | 登録日 | 2022-08-29 | | 公開日 | 2023-09-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8D4N
 
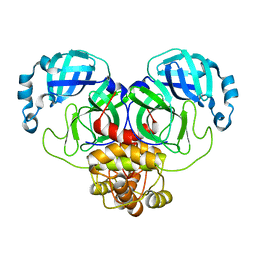 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166Q Mutant | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
8D4J
 
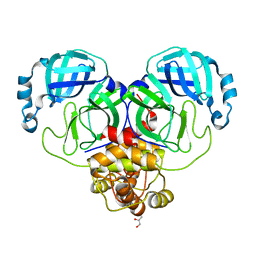 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H172Y Mutant | | 分子名称: | 3C-like proteinase nsp5, GLYCEROL | | 著者 | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | 登録日 | 2022-06-02 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir.
Biorxiv, 2022
|
|
