7E2U
 
 | | Synechocystis GUN4 in complex with phytochrome | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, ... | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2T
 
 | | Synechocystis GUN4 in complex with phycocyanobilin | | Descriptor: | 3-[2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, Ycf53-like protein | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2S
 
 | | Synechocystis GUN4 in complex with biliverdin IXa | | Descriptor: | BILIVERDINE IX ALPHA, GLYCEROL, Ycf53-like protein | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
5ZJL
 
 | |
7CTE
 
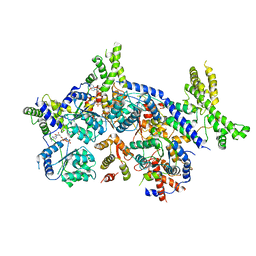 | | Human Origin Recognition Complex, ORC2-5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 2, Origin recognition complex subunit 3, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
7CTF
 
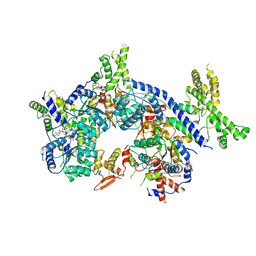 | | Human origin recognition complex 1-5 State II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
6C1W
 
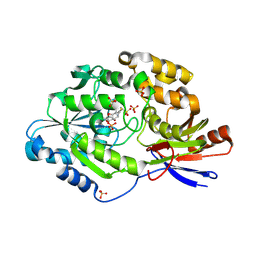 | | A tethered niacin-derived pincer complex with a nickel-carbon or sulfite-carbon bond in lactate racemase | | Descriptor: | (4S)-5-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-4-sulfo-1,4-dihydropyridine-3-carbothioic S-acid, 3-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-5-(sulfanylcarbonyl)pyridin-1-ium, Lactate racemase, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2018-01-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Lactate Racemase Nickel-Pincer Cofactor Operates by a Proton-Coupled Hydride Transfer Mechanism.
Biochemistry, 57, 2018
|
|
7CTG
 
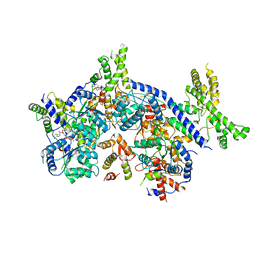 | | Human Origin Recognition Complex, ORC1-5 State I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
6BWR
 
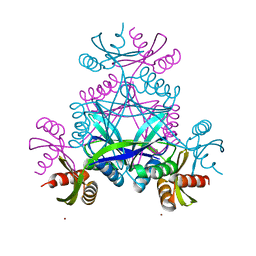 | | LarC2, the C-terminal domain of a cyclometallase involved in the synthesis of the NPN cofactor of lactate racemase, in complex with nickel | | Descriptor: | NICKEL (II) ION, Pyridinium-3,5-bisthiocarboxylic acid mononucleotide nickel insertion protein | | Authors: | Fellner, M, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-12-15 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Biosynthesis of the nickel-pincer nucleotide cofactor of lactate racemase requires a CTP-dependent cyclometallase.
J. Biol. Chem., 293, 2018
|
|
6BWQ
 
 | | LarC2, the C-terminal domain of a cyclometallase involved in the synthesis of the NPN cofactor of lactate racemase, in complex with MnCTP | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Fellner, M, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-12-15 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biosynthesis of the nickel-pincer nucleotide cofactor of lactate racemase requires a CTP-dependent cyclometallase.
J. Biol. Chem., 293, 2018
|
|
6BWO
 
 | | LarC2, the C-terminal domain of a cyclometallase involved in the synthesis of the NPN cofactor of lactate racemase, apo form | | Descriptor: | Pyridinium-3,5-bisthiocarboxylic acid mononucleotide nickel insertion protein | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-12-15 | | Release date: | 2018-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Biosynthesis of the nickel-pincer nucleotide cofactor of lactate racemase requires a CTP-dependent cyclometallase.
J. Biol. Chem., 293, 2018
|
|
6CF3
 
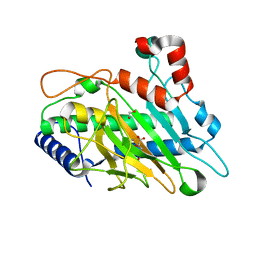 | | Ethylene forming enzyme Y306A variant in complex with manganese and 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-13 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural, Spectroscopic, and Computational Insights from Canavanine-Bound and Two Catalytically Compromised Variants of the Ethylene-Forming Enzyme.
Biochemistry, 2024
|
|
8ZMN
 
 | | Crystal structure of ANTXR1 | | Descriptor: | Anthrax toxin receptor 1, SODIUM ION | | Authors: | Zheng, H, Hu, J, Fu, L, Chen, R. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | X-ray structure of Anthrax toxin receptor 1 APO from Rattus norvegicus
To Be Published
|
|
6B2O
 
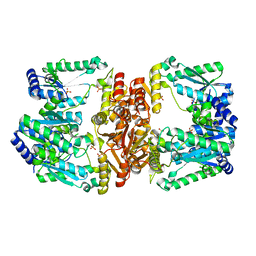 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, C176A variant | | Descriptor: | ATP-utilizing enzyme of the PP-loopsuperfamily, PHOSPHATE ION, SULFATE ION | | Authors: | Fellner, M, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Analysis of the Active Site Cysteine Residue of the Sacrificial Sulfur Insertase LarE from Lactobacillus plantarum.
Biochemistry, 57, 2018
|
|
6B2M
 
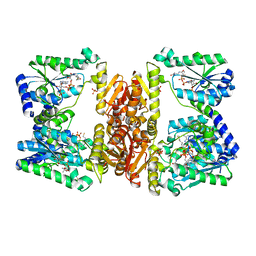 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with coenzyme A | | Descriptor: | ATP-utilizing enzyme of the PP-loopsuperfamily, COENZYME A, PHOSPHATE ION | | Authors: | Fellner, M, Hausinger, R.P, Hu, J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Analysis of the Active Site Cysteine Residue of the Sacrificial Sulfur Insertase LarE from Lactobacillus plantarum.
Biochemistry, 57, 2018
|
|
5TSA
 
 | | Crystal structure of the Zrt-/Irt-like protein from Bordetella bronchiseptica with bound Zn2+ | | Descriptor: | CADMIUM ION, Membrane protein, ZINC ION | | Authors: | Zhang, T, Fellner, M, Sui, D, Liu, J, Hu, J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway.
Sci Adv, 3, 2017
|
|
5TSB
 
 | | Crystal structure of the Zrt-/Irt-like protein from Bordetella bronchiseptica with bound Cd2+ | | Descriptor: | CADMIUM ION, Membrane protein | | Authors: | Zhang, T, Fellner, M, Sui, D, Liu, J, Hu, J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-09-20 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway.
Sci Adv, 3, 2017
|
|
5ULO
 
 | | Crystal Structure of 14-3-3 zeta in Complex with a Serine 124-phosphorylated TBC1D7 peptide | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein zeta/delta, L-PROLINAMIDE, ... | | Authors: | DONG, A, HU, J, MADIGAN, J, WALKER, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-25 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal Structure of 14-3-3 zeta in Complex with a Serine 124-phosphorylated TBC1D7 peptide
to be published
|
|
5V2Z
 
 | | Ethylene forming enzyme in complex with manganese, 2-oxoadipic acid and L-arginine | | Descriptor: | 2-OXOADIPIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
5UDW
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with nickel | | Descriptor: | Lactate racemization operon protein LarE, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UDX
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with zinc | | Descriptor: | Lactate racemization operon protein LarE, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UDT
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Lactate racemization operon protein LarE, PHOSPHATE ION | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UDQ
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, apo form | | Descriptor: | Lactate racemization operon protein LarE, PHOSPHATE ION, SULFATE ION | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UDV
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase, in complex with iron | | Descriptor: | FE (III) ION, Lactate racemization operon protein LarE, PHOSPHATE ION, ... | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UDR
 
 | | LarE, a sulfur transferase involved in synthesis of the cofactor for lactate racemase in complex with nicotinamide mononucleotid NMN | | Descriptor: | ATP-utilizing enzyme of the PP-loopsuperfamily, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MANGANESE (II) ION | | Authors: | Fellner, M, Desguin, B, Hausinger, R.P, Hu, J. | | Deposit date: | 2016-12-28 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
