6UKS
 
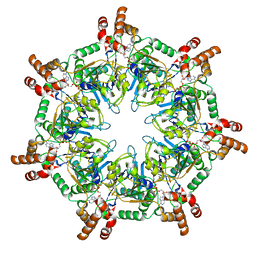 | | ATPgammaS bound mBcs1 | | Descriptor: | MAGNESIUM ION, Mitochondrial chaperone BCS1, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Tang, W.K, Borgnia, M.J, Hsu, A.L, Xia, D. | | Deposit date: | 2019-10-05 | | Release date: | 2020-02-05 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6UKP
 
 | | Apo mBcs1 | | Descriptor: | Mitochondrial chaperone BCS1 | | Authors: | Tang, W.K, Borgnia, M.J, Hsu, A.L, Xia, D. | | Deposit date: | 2019-10-05 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PKW
 
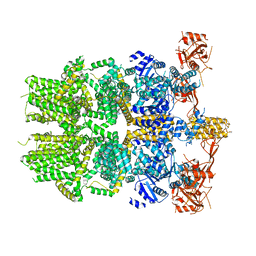 | | Cryo-EM structure of the zebrafish TRPM2 channel in the apo conformation, processed with C2 symmetry (pseudo C4 symmetry) | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
6PKV
 
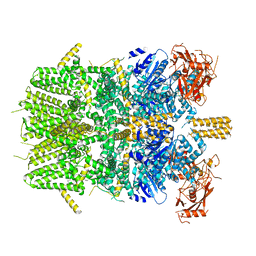 | | Cryo-EM structure of the zebrafish TRPM2 channel in the apo conformation, processed with C4 symmetry | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
6PQP
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist BITC | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-benzylthioformamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
6PQQ
 
 | | Cryo-EM structure of human TRPA1 C621S mutant in the apo state | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily A member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
6PKX
 
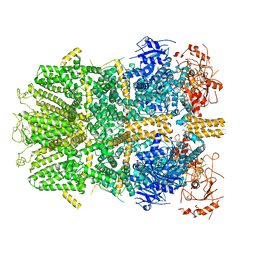 | | Cryo-EM structure of the zebrafish TRPM2 channel in the presence of ADPR and Ca2+ | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 2, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
6PQO
 
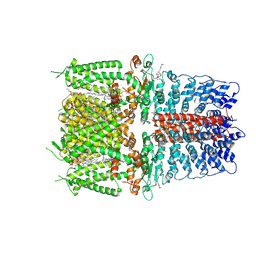 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist JT010 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-N-(3-methoxypropyl)acetamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
6OT5
 
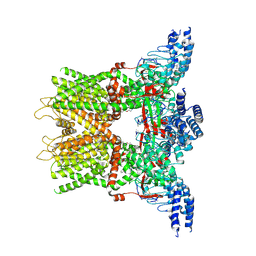 | | Structure of the TRPV3 K169A sensitized mutant in the presence of 2-APB at 3.6 A resolution | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 3,Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Borschel, W.F, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Regulatory switch at the cytoplasmic interface controls TRPV channel gating.
Elife, 8, 2019
|
|
6OO4
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in amphipol resolved to 3.3 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
6OO3
 
 | | Cryo-EM structure of the C4-symmetric TRPV2/RTx complex in amphipol resolved to 2.9 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
6OF4
 
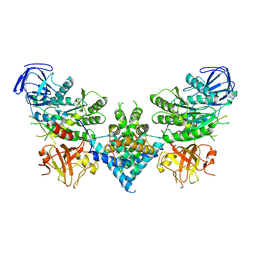 | | Precursor ribosomal RNA processing complex, apo-state. | | Descriptor: | CLP1_P domain-containing protein, Ribonuclease | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OT2
 
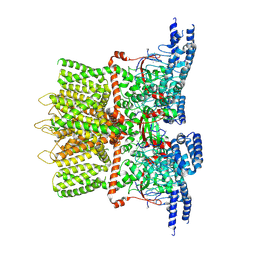 | | Structure of the TRPV3 K169A sensitized mutant in apo form at 4.1 A resolution | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Borschel, W.F, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Regulatory switch at the cytoplasmic interface controls TRPV channel gating.
Elife, 8, 2019
|
|
7TU6
 
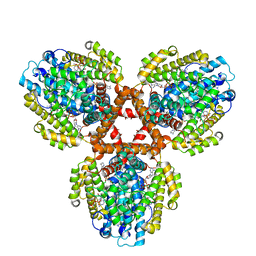 | | Structure of the L. blandensis dGTPase bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, dGTP triphosphohydrolase | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU7
 
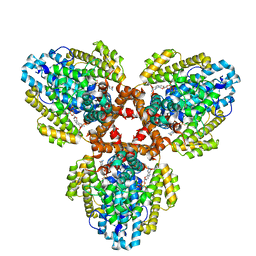 | | Structure of the L. blandensis dGTPase H125A mutant bound to dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, dGTP triphosphohydrolase | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU8
 
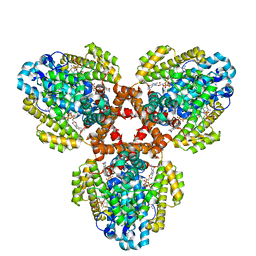 | | Structure of the L. blandensis dGTPase H125A mutant bound to dGTP and dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7TU5
 
 | | Structure of the L. blandensis dGTPase in the apo form | | Descriptor: | MAGNESIUM ION, dGTP triphosphohydrolase | | Authors: | Klemm, B.P, Sikkema, A.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution structures of the SAMHD1 dGTPase homolog from Leeuwenhoekiella blandensis reveal a novel mechanism of allosteric activation by dATP.
J.Biol.Chem., 298, 2022
|
|
7U65
 
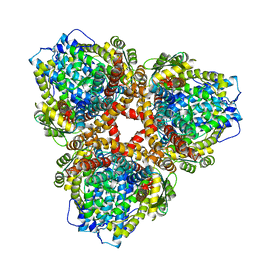 | | Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, Inhibitor of dGTPase | | Authors: | Klemm, B.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism by which T7 bacteriophage protein Gp1.2 inhibits Escherichia coli dGTPase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U67
 
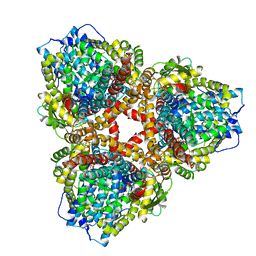 | | Structure of E. coli dGTPase bound to T7 bacteriophage protein Gp1.2 and GTP | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, GUANOSINE-5'-TRIPHOSPHATE, Inhibitor of dGTPase, ... | | Authors: | Klemm, B.P, Hsu, A.L, Borgnia, M.J, Schaaper, R.M. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism by which T7 bacteriophage protein Gp1.2 inhibits Escherichia coli dGTPase.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6MAT
 
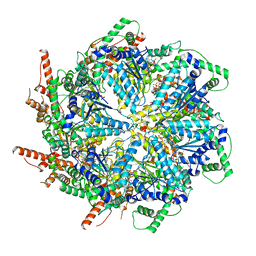 | | Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Rix7 mutant, unknown protein | | Authors: | Lo, Y.H, Sobhany, M, Hsu, A.L, Ford, B.L, Krahn, J.M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2018-08-28 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the essential ribosome assembly AAA-ATPase Rix7.
Nat Commun, 10, 2019
|
|
6NR4
 
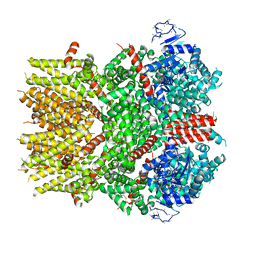 | | Cryo-EM structure of the TRPM8 ion channel with low occupancy icilin, PI(4,5)P2, and calcium | | Descriptor: | Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6NR2
 
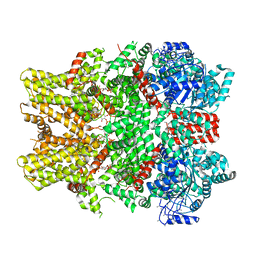 | | Cryo-EM structure of the TRPM8 ion channel in complex with the menthol analog WS-12 and PI(4,5)P2 | | Descriptor: | (1R,2S,5R)-N-(4-methoxyphenyl)-5-methyl-2-(propan-2-yl)cyclohexane-1-carboxamide, (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6NR3
 
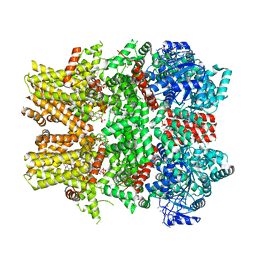 | | Cryo-EM structure of the TRPM8 ion channel in complex with high occupancy icilin, PI(4,5)P2, and calcium | | Descriptor: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, CALCIUM ION, Icilin, ... | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6OF2
 
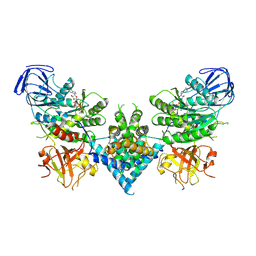 | | Precursor ribosomal RNA processing complex, State 2. | | Descriptor: | CLP1_P domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OF3
 
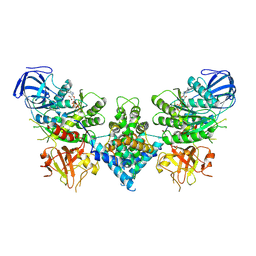 | | Precursor ribosomal RNA processing complex, State 1. | | Descriptor: | CLP1_P domain-containing protein, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
