3W53
 
 | |
5Y6G
 
 | | PilZ domain with c-di-GMP of YcgR from Escherichia coli | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | 著者 | Hou, Y.J, Wang, D.C, Li, D.F. | | 登録日 | 2017-08-11 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
5Y6H
 
 | |
5Y6F
 
 | | Crystal structure of YcgR in complex with c-di-GMP from Escherichia coli | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | 著者 | Hou, Y.J, Wang, D.C, Li, D.F. | | 登録日 | 2017-08-11 | | 公開日 | 2018-07-18 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
8JYW
 
 | |
8JYZ
 
 | | Cryo-EM structure of RCD-1 pore from Neurospora crassa | | 分子名称: | Gasdermin-like protein rcd-1-1, Gasdermin-like protein rcd-1-2 | | 著者 | Hou, Y.J, Sun, Q, Li, Y, Ding, J. | | 登録日 | 2023-07-04 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.63 Å) | | 主引用文献 | Cleavage-independent activation of ancient eukaryotic gasdermins and structural mechanisms.
Science, 384, 2024
|
|
8GTN
 
 | |
7WR1
 
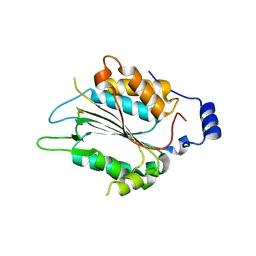 | | P32 of caspase-4 C258A mutant in complex with OspC3 C-terminal ankyrin-repeat domain | | 分子名称: | Caspase-4, OspC3 | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR4
 
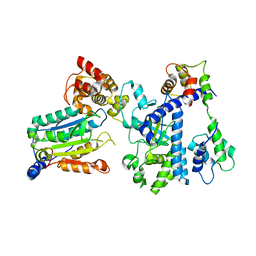 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | 分子名称: | Calmodulin-1, Caspase-4, OspC3 | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR0
 
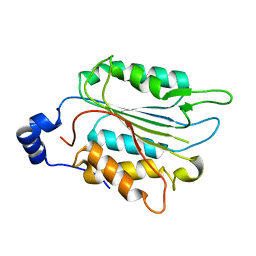 | | P32 of caspase-4 C258A mutant | | 分子名称: | Caspase-4 | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR6
 
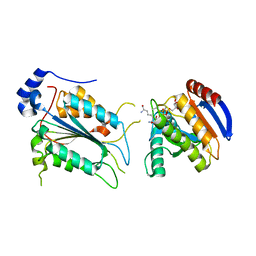 | | Crystal structure of ADP-riboxanated caspase-4 in complex with Af1521 | | 分子名称: | ADP-ribose glycohydrolase AF_1521, Caspase-4, [[(3~{a}~{S},5~{R},6~{R},6~{a}~{R})-2-azanylidene-3-[(4~{R})-4-azanyl-5-oxidanylidene-pentyl]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxazol-5-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR3
 
 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | 分子名称: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR2
 
 | |
7WR5
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex binding with 2'-aF-NAD+ | | 分子名称: | Calmodulin-1, Caspase-4, OspC3, ... | | 著者 | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | 登録日 | 2022-01-26 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3AQS
 
 | | Crystal structure of RolR (NCGL1110) without ligand | | 分子名称: | Bacterial regulatory proteins, tetR family | | 著者 | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | 登録日 | 2010-11-18 | | 公開日 | 2011-07-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
3AQT
 
 | | CRYSTAL STRUCTURE OF RolR (NCGL1110) complex WITH ligand RESORCINOL | | 分子名称: | Bacterial regulatory proteins, tetR family, RESORCINOL | | 著者 | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | 登録日 | 2010-11-18 | | 公開日 | 2011-07-06 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
3VSG
 
 | | Crystal structure of iron free 1,6-APD, 2-Animophenol-1,6-Dioxygenase | | 分子名称: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit | | 著者 | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | 登録日 | 2012-04-25 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSI
 
 | | Crystal structure of native 1,6-APD (2-Animophenol-1,6-dioxygenase) complex with 4-Nitrocatechol | | 分子名称: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit, 4-NITROCATECHOL, ... | | 著者 | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | 登録日 | 2012-04-25 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSJ
 
 | | Crystal structure of 1,6-APD (2-ANIMOPHENOL-1,6-DIOXYGENASE) complexed with intermediate products | | 分子名称: | (2Z,4Z)-2-imino-6-oxohex-4-enoic acid, (3E)-3-iminooxepin-2(3H)-one, 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, ... | | 著者 | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | 登録日 | 2012-04-25 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSH
 
 | | Crystal structure of native 1,6-APD (with Iron), 2-Animophenol-1,6-Dioxygenase | | 分子名称: | 2-amino-5-chlorophenol 1,6-dioxygenase alpha subunit, 2-amino-5-chlorophenol 1,6-dioxygenase beta subunit, FE (II) ION | | 著者 | Li, D.F, Hou, Y.J, Hu, Y, Wang, D.C, Liu, W. | | 登録日 | 2012-04-25 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of aminophenol dioxygenase in complex with intermediate, product and inhibitor
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WMS
 
 | | The crystal structure of Y195I mutant alpha-cyclodextrin glycosyltransferase from Paenibacillus macerans | | 分子名称: | Alpha-cyclodextrin glucanotransferase, CALCIUM ION | | 著者 | Xie, T, Hou, Y.J, Li, D.F, Yue, Y, Qian, S.J, Chao, Y.P. | | 登録日 | 2013-11-24 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of a mutant Y195I alpha-cyclodextrin glycosyltransferase with switched product specificity from alpha-cyclodextrin to beta-/ gamma-cyclodextrin
J.Biotechnol., 182-183, 2014
|
|
6LLB
 
 | | Crystal structure of mpy-RNase J (mutant S247A), an archaeal RNase J from Methanolobus psychrophilus R15, in complex with 6 nt RNA | | 分子名称: | MPY-RNase J, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, ... | | 著者 | Li, D.F, Hou, Y.J, Guo, L. | | 登録日 | 2019-12-22 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A newly identified duplex RNA unwinding activity of archaeal RNase J depends on processive exoribonucleolysis coupled steric occlusion by its structural archaeal loops.
Rna Biol., 17, 2020
|
|
5Z2L
 
 | | Crystal structure of BdcA in complex with NADPH | | 分子名称: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Cyclic-di-GMP-binding biofilm dispersal mediator protein, ... | | 著者 | Yang, W.S, Hou, Y.J, Li, D.F, Wang, D.C. | | 登録日 | 2018-01-03 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A potential substrate binding pocket of BdcA plays a critical role in NADPH recognition and biofilm dispersal
Biochem. Biophys. Res. Commun., 497, 2018
|
|
8JYV
 
 | |
8JYX
 
 | |
