3RIG
 
 | |
3RIY
 
 | |
6IQL
 
 | | Crystal structure of dopamine receptor D4 bound to the subtype-selective ligand, L745870 | | Descriptor: | 3-{[4-(4-chlorophenyl)piperazin-1-yl]methyl}-1H-pyrrolo[2,3-b]pyridine, D(4) dopamine receptor,Soluble cytochrome b562,D(4) dopamine receptor | | Authors: | Zhou, Y, Cao, C, Zhang, X.C. | | Deposit date: | 2018-11-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of dopamine receptor D4 bound to the subtype selective ligand, L745870.
Elife, 8, 2019
|
|
6KIM
 
 | |
4F4U
 
 | |
4F56
 
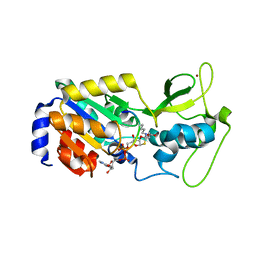 | | The bicyclic intermediate structure provides insights into the desuccinylation mechanism of SIRT5 | | Descriptor: | 3-[(2R,3aR,5R,6R,6aR)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-2,6-dihydroxytetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]propanoic acid, NAD-dependent lysine demalonylase and desuccinylase sirtuin-5, mitochondrial, ... | | Authors: | Zhou, Y, Hao, Q. | | Deposit date: | 2012-05-11 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Bicyclic Intermediate Structure Provides Insights into the Desuccinylation Mechanism of Human Sirtuin 5 (SIRT5)
J.Biol.Chem., 287, 2012
|
|
2K74
 
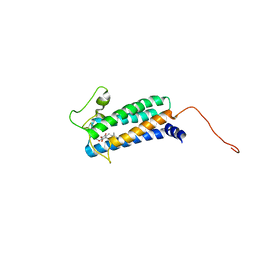 | | Solution NMR structure of DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, UBIQUINONE-2 | | Authors: | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
2K73
 
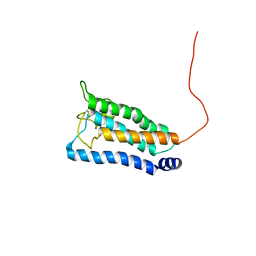 | | Solution NMR structure of integral membrane protein DsbB | | Descriptor: | Disulfide bond formation protein B | | Authors: | Zhou, Y, Cierpicki, T, Flores Jimenez, R.H, Lukasik, S.M, Ellena, J.F, Cafiso, D.S, Kadokura, H, Beckwith, J, Bushweller, J.H. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation.
Mol.Cell, 31, 2008
|
|
3U31
 
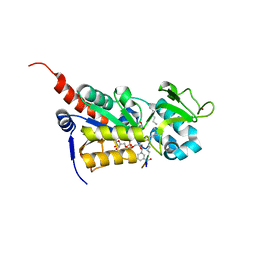 | |
3U3D
 
 | |
2HVJ
 
 | | Crystal structure of KcsA-Fab-TBA complex in low K+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel.
J.Mol.Biol., 366, 2007
|
|
2HVK
 
 | | crystal structure of the KcsA-Fab-TBA complex in high K+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, Antibody Fab heavy chain, Antibody Fab light chain, ... | | Authors: | Zhou, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel.
J.Mol.Biol., 366, 2007
|
|
8GUL
 
 | |
8GUM
 
 | |
1R3J
 
 | | potassium channel KcsA-Fab complex in high concentration of Tl+ | | Descriptor: | Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, DIACYL GLYCEROL, ... | | Authors: | Zhou, Y, MacKinnon, R. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The occupancy of ions in the K+ selectivity filter: Charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates
J.Mol.Biol., 333, 2003
|
|
1R3L
 
 | | potassium channel KcsA-Fab complex in Cs+ | | Descriptor: | Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, CESIUM ION, ... | | Authors: | Zhou, Y, MacKinnon, R. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The occupancy of ions in the K+ selectivity filter: Charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates
J.Mol.Biol., 333, 2003
|
|
1R3K
 
 | | potassium channel KcsA-Fab complex in low concentration of Tl+ | | Descriptor: | Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, DIACYL GLYCEROL, ... | | Authors: | Zhou, Y, MacKinnon, R. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The occupancy of ions in the K+ selectivity filter: Charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates
J.Mol.Biol., 333, 2003
|
|
1R3I
 
 | | potassium channel KcsA-Fab complex in Rb+ | | Descriptor: | Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, DIACYL GLYCEROL, ... | | Authors: | Zhou, Y, MacKinnon, R. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The occupancy of ions in the K+ selectivity filter: Charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates
J.Mol.Biol., 333, 2003
|
|
7SS9
 
 | | Late translocation intermediate with EF-G partially dissociated (Structure V) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Carbone, C.E, Loveland, A.B, Gamper, H.B, Hou, Y, Korostelev, A.A. | | Deposit date: | 2021-11-10 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP.
Nat Commun, 12, 2021
|
|
6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Descriptor: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Authors: | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
8W6P
 
 | | Crystal structure of dimeric murine SMPDL3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Zhang, C, Liu, P, Fan, S, Hou, Y. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
8VZ0
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-400 | | Descriptor: | (1S,2R,4S)-N-(cyclopropylmethyl)-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VZ1
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-409 | | Descriptor: | 4,4'-[(1S,4S,5R)-5-(6-methoxy-3,4-dihydroquinoline-1(2H)-sulfonyl)-7-oxabicyclo[2.2.1]hept-2-ene-2,3-diyl]diphenol, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VYT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-411 | | Descriptor: | 4,4'-[(1R,4R,5S)-5-(2,3-dihydro-1H-indole-1-sulfonyl)-7-oxabicyclo[2.2.1]hept-2-ene-2,3-diyl]diphenol, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-09 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VZP
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (L372S, L536S) in complex with k-403 | | Descriptor: | (1S,2R,4S)-N-(2-hydroxyethyl)-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor | | Authors: | Min, C.K, Nwachukwu, J.C, Hou, Y, Russo, R.J, Papa, A, Min, J, Peng, R, Kim, S.H, Ziegler, Y, Rangarajan, E.S, Izard, T, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Asymmetric allostery in estrogen receptor-alpha homodimers drives responses to the ensemble of estrogens in the hormonal milieu.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
