6VAM
 
 | |
6VAK
 
 | | Cryo-EM structure of human CALHM2 | | Descriptor: | Calcium homeostasis modulator protein 2 | | Authors: | Syrjanen, J.L, Chou, T.H, Furukawa, H. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure and assembly of calcium homeostasis modulator proteins.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3R6J
 
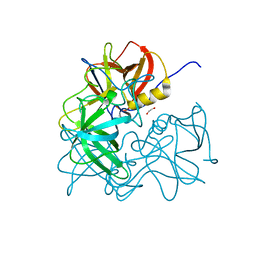 | | Crystal Structure of the Capsid P Domain from Norwalk Virus Strain Hiroshima/1999 | | Descriptor: | 1,2-ETHANEDIOL, VP1 protein | | Authors: | Hansman, G.S, Biertumpfel, C, McLellan, J.S, Georgiev, I, Chen, L, Zhou, T, Katayama, K, Kwong, P.D. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
7L5X
 
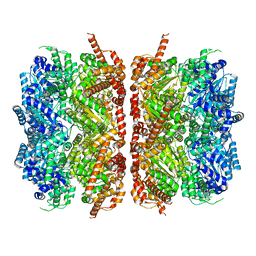 | | p97-R155H mutant dodecamer II | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7L5W
 
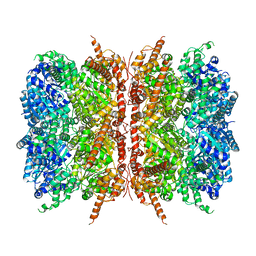 | | p97-R155H mutant dodecamer I | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
4YFL
 
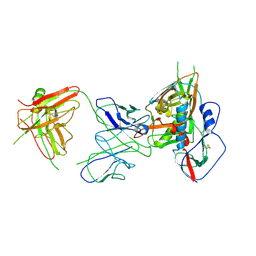 | | Crystal structure of VH1-46 germline-derived CD4-binding site-directed antibody 1B2530 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 1B2530 Light chain, 1B2530 heavy chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Acharya, P, Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2015-02-25 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.387 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
7UEB
 
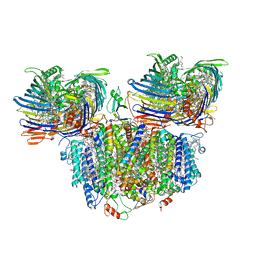 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO2) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
7UEA
 
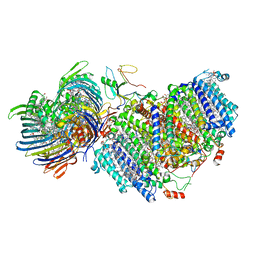 | | Photosynthetic assembly of Chlorobaculum tepidum (RC-FMO1) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Puskar, R, Truong, C.D, Swain, K, Li, S, Cheng, K.-W, Wang, T.Y, Poh, Y.-P, Liu, H, Chou, T.-F, Nannenga, B, Chiu, P.-L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular asymmetry of a photosynthetic supercomplex from green sulfur bacteria.
Nat Commun, 13, 2022
|
|
3R6K
 
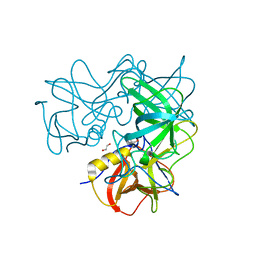 | | Crystal Structure of the Capsid P Domain from Norwalk Virus Strain Hiroshima/1999 in complex with HBGA type B (triglycan) | | Descriptor: | 1,2-ETHANEDIOL, VP1 protein, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Hansman, G.S, Biertumpfel, C, McLellan, J.S, Georgiev, I, Chen, L, Zhou, T, Katayama, K, Kwong, P.D. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of GII.10 and GII.12 norovirus protruding domains in complex with histo-blood group antigens reveal details for a potential site of vulnerability.
J.Virol., 85, 2011
|
|
6USV
 
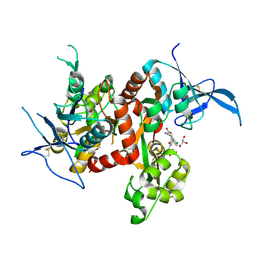 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and SDZ 220-040 | | Descriptor: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, GLYCEROL, GLYCINE, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, h. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6ZR0
 
 | |
6ZQR
 
 | |
6ZQY
 
 | |
6ZR4
 
 | |
6ZR3
 
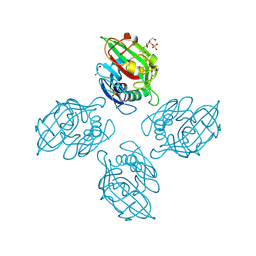 | |
6ZQX
 
 | | Crystal structure of tetrameric fibrinogen-like recognition domain of FIBCD1 with N,N'-diacetyl chitobiose ligand bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CALCIUM ION, ... | | Authors: | Shrive, A.K, Greenhough, T.J, Williams, H.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of human immune protein FIBCD1 suggest an extended binding site compatible with recognition of pathogen associated carbohydrate motifs
J.Biol.Chem., 2023
|
|
6USU
 
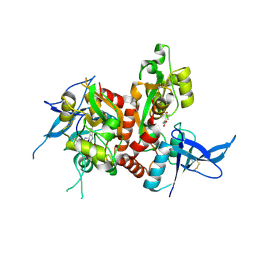 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with L689,560 and glutamate | | Descriptor: | (2R,4S)-5,7-dichloro-4-[(phenylcarbamoyl)amino]-1,2,3,4-tetrahydroquinoline-2-carboxylic acid, GLUTAMIC ACID, Glutamate receptor ionotropic, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, H. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
4ZJM
 
 | | Crystal Structure of Mycobacterium tuberculosis LpqH (Rv3763) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein LpqH, ... | | Authors: | Arbing, M.A, Chan, S, Kuo, E, Harris, L.R, Zhou, T.T, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis LpqH (Rv3763)
To Be Published
|
|
3JWO
 
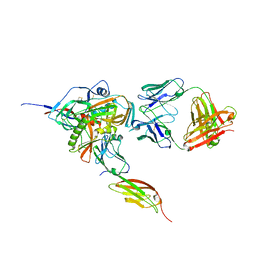 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D Heavy CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Huang, C.C, Kwon, Y.D, Zhou, T, Robinson, J.E, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JWD
 
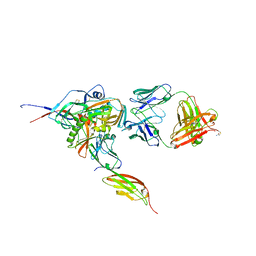 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D HEAVY CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Ban, Y.A, Chen, L, Huang, C.C, Kong, L, Kwon, Y.D, Stuckey, J, Zhou, T, Robinson, J.E, Schief, W.R, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | Authors: | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-05-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IDX
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab b13 heavy chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IDY
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab b13 heavy chain, Fab b13 light chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
4GZR
 
 | | Crystal structure of the Mycobacterium tuberculosis H37Rv EsxOP (Rv2346c-Rv2347c) complex in space group C2221 | | Descriptor: | ESAT-6-like protein 6, ESAT-6-like protein 7, SULFATE ION | | Authors: | Arbing, M.A, Chan, S, He, Q, Harris, L, Zhou, T.T, Kuo, E, Ahn, C.J, Eisenberg, D, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2012-09-06 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
4PM4
 
 | | Structure of a putative periplasmic iron siderophore binding protein (Rv0265c) from Mycobacterium tuberculosis H37Rv | | Descriptor: | CHLORIDE ION, Iron complex transporter substrate-binding protein, SULFATE ION | | Authors: | Arbing, M.A, Chan, S, Tran, N, Kuo, E, Lu, J, Harris, L.R, Zhou, T.T, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-05-20 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a putative periplasmic iron siderophore binding protein (Rv0265c) from Mycobacterium tuberculosis H37Rv
To Be Published
|
|
