7END
 
 | |
7EN9
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-02 | | Descriptor: | 3C-like proteinase, 5-bromanyl-~{N}-methyl-3-nitro-2-[(4~{R},5~{S})-2-(7-oxidanylisoquinolin-4-yl)carbonyl-4-phenyl-2,7-diazaspiro[4.4]nonan-7-yl]benzamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7EN8
 
 | | Crystal structure of SARS-CoV-2 3CLpro in complex with the non-covalent inhibitor WU-04 | | Descriptor: | 3C-like proteinase, GLYCEROL, ~{N}-[(1~{S},2~{R})-2-[[4-bromanyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]isoquinoline-4-carboxamide | | Authors: | Hou, N, Peng, C, Hu, Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro.
Acs Cent.Sci., 9, 2023
|
|
7ENE
 
 | |
8X6Z
 
 | |
8I37
 
 | | Helicobacter pylori G6PDH | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Zhou, N, Gao, L.Z. | | Deposit date: | 2023-01-16 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Inorganic polysulfide kills Helicobacter pylori by inactivating G6PDH via cysteine polysulfidation
To Be Published
|
|
8DK4
 
 | | Peroxisome proliferator-activated receptor gamma in complex with VSP-51-2 | | Descriptor: | 5-(benzylcarbamoyl)-1-[(4-chloro-3-fluorophenyl)methyl]-1H-indole-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Ma, L, Zhou, X.E, Suino-Powell, K, Hou, N, Zhou, Z, Luo, J, Xu, H.E, Yi, W. | | Deposit date: | 2022-07-02 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of VSP-51-2 as the Novel and Safe PPAR gamma Modulator: Structure-Based Design, Biological Validation and Crystal Analysis
To Be Published
|
|
5H0S
 
 | | EM Structure of VP1A and VP1B | | Descriptor: | VP1 | | Authors: | Li, X, Zhou, N, Xu, B, Chen, W, Zhu, B, Wang, X, Wang, J, Liu, H, Cheng, L. | | Deposit date: | 2016-10-06 | | Release date: | 2017-01-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
5H0R
 
 | | RNA dependent RNA polymerase ,vp4,dsRNA | | Descriptor: | RNA (42-MER), RNA-dependent RNA polymerase, VP4 protein | | Authors: | Li, X, Zhou, N, Chen, W, Zhu, B, Wang, X, Xu, B, Wang, J, Liu, H, Cheng, L. | | Deposit date: | 2016-10-06 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
7CZA
 
 | |
7NSU
 
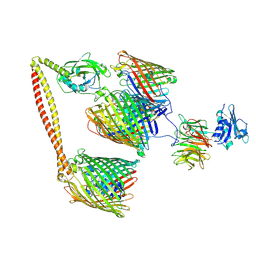 | | ColicinE9 intact translocation complex | | Descriptor: | Colicin-E9, Outer membrane protein F, Tol-Pal system protein TolB, ... | | Authors: | Webby, M.N, Kleanthous, C, Lukoyanova, N, Housden, N.G. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-11 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Porin threading drives receptor disengagement and establishes active colicin transport through Escherichia coli OmpF.
Embo J., 40, 2021
|
|
7NST
 
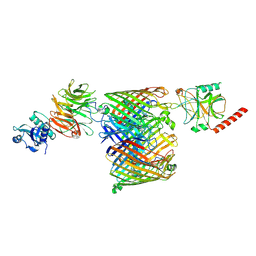 | | ColicinE9 partial translocation complex | | Descriptor: | Colicin-E9, Outer membrane protein F, Tol-Pal system protein TolB | | Authors: | Webby, M.N, Kleanthous, C, Lukoyanova, N, Housden, N.G. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Porin threading drives receptor disengagement and establishes active colicin transport through Escherichia coli OmpF.
Embo J., 40, 2021
|
|
7NG9
 
 | |
7NG8
 
 | |
7NNA
 
 | |
8WWU
 
 | | 1-naphthylamine GS in complex with AMP PNP | | Descriptor: | Glutamine synthetase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Zhang, S.T, Zhou, N.Y. | | Deposit date: | 2023-10-26 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of the 1-naphthylamine biodegradation pathway reveals a glutamine synthetase-like protein that catalyzes 1-naphthylamine glutamylation
To Be Published
|
|
8WWV
 
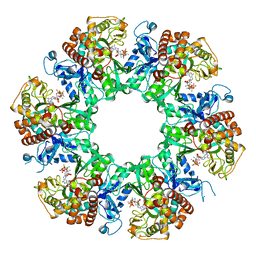 | | 1-naphthylamine GS in complex with ADP and MetSox-P | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Zhang, S.T, Zhou, N.Y. | | Deposit date: | 2023-10-26 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the 1-naphthylamine biodegradation pathway reveals a glutamine synthetase-like protein that catalyzes 1-naphthylamine glutamylation
To Be Published
|
|
8X6V
 
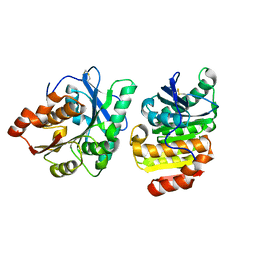 | | Crystal structure of GlacPETase | | Descriptor: | GlacPETase | | Authors: | Qi, X, Zhou, N.Y. | | Deposit date: | 2023-11-22 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The unique salt bridge network in GlacPETase: a key to its stability.
Appl.Environ.Microbiol., 90, 2024
|
|
6TZV
 
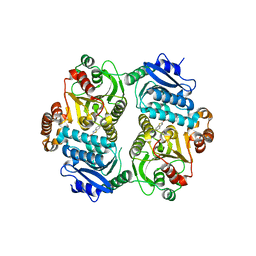 | | Crystal Structure of the carboxyltransferase subunit of ACC (AccD6) in complex with inhibitor Phenyl-Cyclodiaone from Mycobacterium tuberculosis | | Descriptor: | 4'-[(6-chloro-1,3-benzothiazol-2-yl)oxy]-6-hydroxy-4,4-dimethyl-4,5-dihydro[1,1'-biphenyl]-2(3H)-one, Probable propionyl-CoA carboxylase beta chain 6 | | Authors: | Reddy, M.C.M, Zhou, N, Sacchettini, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-08-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Novel herbicidal derivatives that inhibit carboxyltransferase subunit of the acetyl-CoA carboxylase in Mycobacterium tuberculosis
To Be Published
|
|
7ZQ8
 
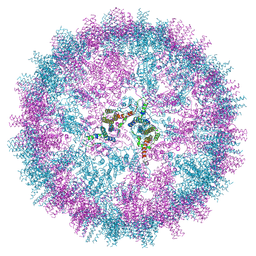 | | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR (T=4 VLP) | | Descriptor: | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR | | Authors: | Kingston, N.J, Grehan, K, Snowden, J.S, Alzahrani, J, Ranson, N.A, Rowlands, D.J, Stonehouse, N.J. | | Deposit date: | 2022-04-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | VelcroVax: a "Bolt-On" Vaccine Platform for Glycoprotein Display.
Msphere, 8, 2023
|
|
7ZQA
 
 | | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR (T=3* VLP) | | Descriptor: | VelcroVax tandem HBcAg with SUMO-Affimer inserted at MIR | | Authors: | Kingston, N.J, Grehan, K, Snowden, J.S, Alzahrani, J, Ranson, N.A, Rowlands, D.J, Stonehouse, N.J. | | Deposit date: | 2022-04-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | VelcroVax: a "Bolt-On" Vaccine Platform for Glycoprotein Display.
Msphere, 8, 2023
|
|
6S6L
 
 | | Cryo-EM structure of murine norovirus (MNV-1) | | Descriptor: | Capsid protein | | Authors: | Snowden, J.S, Hurdiss, D.L, Adeyemi, O.O, Ranson, N.A, Herod, M.R, Stonehouse, N.J. | | Deposit date: | 2019-07-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Dynamics in the murine norovirus capsid revealed by high-resolution cryo-EM.
Plos Biol., 18, 2020
|
|
6LR1
 
 | |
8C6D
 
 | | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate. | | Descriptor: | (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol, Genome polyprotein, Genome polyprotein (Fragment) | | Authors: | Kingston, N.J, Snowden, J.S, Stonehouse, N.J, Rowlands, D.J, Hogle, J.M. | | Deposit date: | 2023-01-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate.
Biorxiv, 2023
|
|
7BC4
 
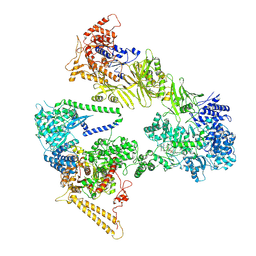 | | Cryo-EM structure of fatty acid synthase (FAS) from Pichia pastoris | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta | | Authors: | Snowden, J.S, Alzahrani, J, Sherry, L, Stacey, M, Rowlands, D.J, Ranson, N.A, Stonehouse, N.J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Pichia pastoris fatty acid synthase.
Sci Rep, 11, 2021
|
|
