7YSO
 
 | |
7V2T
 
 | | The complex structure of SoBcmB and its natural precursor 2 | | Descriptor: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Zhou, J.H, Wu, L. | | Deposit date: | 2021-08-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.20005679 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
8VPG
 
 | |
8VPI
 
 | |
8VPH
 
 | |
4V9L
 
 | | 70S Ribosome translocation intermediate FA-3.6A containing elongation factor EFG/FUSIDIC ACID/GDP, mRNA, and tRNA bound in the pe*/E state. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhou, J, Lancaster, L, Donohue, J.P, Noller, H.F. | | Deposit date: | 2013-04-24 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation.
Science, 340, 2013
|
|
4V9K
 
 | | 70S ribosome translocation intermediate GDPNP-I containing elongation factor EFG/GDPNP, mRNA, and tRNA bound in the pe*/E state. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhou, J, Lancaster, L, Donohue, J.P, Noller, H.F. | | Deposit date: | 2013-04-24 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation.
Science, 340, 2013
|
|
6FHY
 
 | | Photorhabdus asymbiotica lectin (PHL) in complex with synthetic C-fucoside | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{S})-2-[[(2~{S},4~{R},6~{R})-2-(hydroxymethyl)-6-oxidanyl-oxan-4-yl]methyl]-6-methyl-oxane-3,4,5-triol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Houser, J, Jancarikova, G, Wimmerova, M. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fucosylated inhibitors of recently identified bangle lectin from Photorhabdus asymbiotica.
Sci Rep, 9, 2019
|
|
6FLU
 
 | | Photorhabdus asymbiotica lectin (PHL) in complex with synthetic C-fucoside | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{S})-2-[[(2~{R},3~{S},4~{R},6~{S})-2-(hydroxymethyl)-6-methoxy-3-oxidanyl-oxan-4-yl]methyl]-6-methyl-oxane-3,4,5-triol, CHLORIDE ION, Photorhabdus asymbitoca lectin PHL, ... | | Authors: | Houser, J, Jancarikova, G, Wimmerova, M. | | Deposit date: | 2018-01-28 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Fucosylated inhibitors of recently identified bangle lectin from Photorhabdus asymbiotica.
Sci Rep, 9, 2019
|
|
6FPD
 
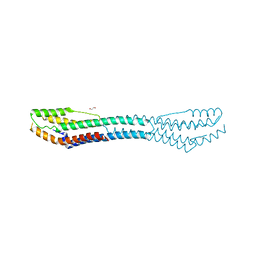 | | AB21 protein from Agaricus bisporus | | Descriptor: | 1,2-ETHANEDIOL, Protein AB21, SODIUM ION | | Authors: | Houser, J, Demo, G, Komarek, J, Wimmerova, M. | | Deposit date: | 2018-02-09 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and properties of AB21, a novel Agaricus bisporus protein with structural relation to bacterial pore-forming toxins.
Proteins, 86, 2018
|
|
8FS1
 
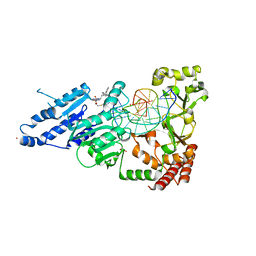 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11a (YD905) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{2-[N'-(cyclohexylmethyl)carbamimidamido]ethyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
8FS2
 
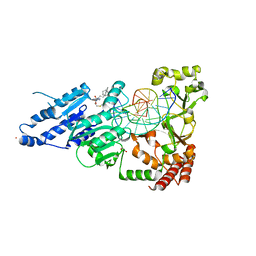 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11b (YD907) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{3-[N'-(cyclohexylmethyl)carbamimidamido]propyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
6FHX
 
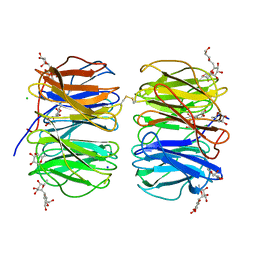 | | Photorhabdus asymbiotica lectin (PHL) in complex with synthetic C-fucoside | | Descriptor: | (2~{S},3~{S},4~{R},5~{S},6~{S})-2-[[(2~{R},3~{S},4~{R},6~{R})-2-(hydroxymethyl)-3,6-bis(oxidanyl)oxan-4-yl]methyl]-6-methyl-oxane-3,4,5-triol, CHLORIDE ION, Lectin PHL, ... | | Authors: | Houser, J, Jancarikova, G, Wimmerova, M. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Fucosylated inhibitors of recently identified bangle lectin from Photorhabdus asymbiotica.
Sci Rep, 9, 2019
|
|
6F5W
 
 | | Photorhabdus asymbiotica lectin (PHL) in complex with propargyl-fucoside | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, SODIUM ION, ... | | Authors: | Houser, J, Jancarikova, G, Wimmerova, M, Csavas, M, Borbas, A, Herczeg, M, Fujdiarova, E, Kover, E.K. | | Deposit date: | 2017-12-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Synthesis of alpha-l-Fucopyranoside-Presenting Glycoclusters and Investigation of Their Interaction with Photorhabdus asymbiotica Lectin (PHL).
Chemistry, 24, 2018
|
|
7OYJ
 
 | |
6GKE
 
 | | Aleuria aurantia lectin AAL N224Q mutant in complex with Fucalpha1-6GlcNAc | | Descriptor: | 1,2-ETHANEDIOL, Fucose-specific lectin, GLYCEROL, ... | | Authors: | Houser, J, Wimmerova, M, Herrera, H, Mehta, A.S. | | Deposit date: | 2018-05-19 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural dynamics and applications of a fucose-binding lectin with enhanced binding to alpha-1,6-linked core fucosylation
To Be Published
|
|
8CY5
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 39 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-{3-[1-(tert-butoxycarbonyl)piperidin-4-yl]propyl}adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY3
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-[3-(4-aminophenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY1
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 19 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(5-phenylpentyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXZ
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-(3-Phenylpropyl)adenosine (Compound 14) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(3-phenylpropyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY4
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 16 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-[3-(4-hydroxyphenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY0
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor MC4756 (Compound 178) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(4-phenylbutyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
3LWO
 
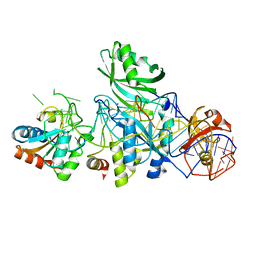 | | Structure of H/ACA RNP bound to a substrate RNA containing 5BrU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(5BU)P*GP*CP*GP*GP*UP*UP*U)-3', 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.855 Å) | | Cite: | Glycosidic bond conformation preference plays a pivotal role in catalysis of RNA pseudouridylation: a combined simulation and structural study.
J.Mol.Biol., 401, 2010
|
|
3LWP
 
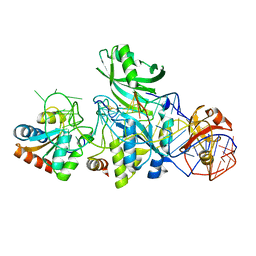 | | Structure of H/ACA RNP bound to a substrate RNA containing 5BrdU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(BRU)P*GP*CP*GP*GP*UP*UP*U)-3, 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glycosidic bond conformation preference plays a pivotal role in catalysis of RNA pseudouridylation: a combined simulation and structural study.
J.Mol.Biol., 401, 2010
|
|
3LWR
 
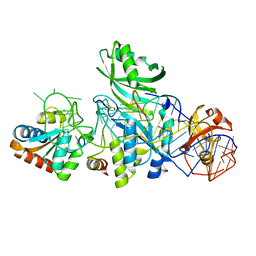 | | Structure of H/ACA RNP bound to a substrate RNA containing 4SU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(4SU)P*GP*CP*GP*GP*UP*UP*U)-3', H/ACA RNA, Large ribosomal subunit protein eL8, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Functional and Structural Impact of Target Uridine Substitutions on the H/ACA Ribonucleoprotein Particle Pseudouridine Synthase .
Biochemistry, 49, 2010
|
|
