5E8I
 
 | |
4Z2Y
 
 | | Crystal structure of methyltransferase CalO6 | | Descriptor: | CalO6, MERCURY (II) ION | | Authors: | Hou, C, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of O-methyltransferase CalO6 from the calicheamicin biosynthetic pathway: a case of challenging structure determination at low resolution.
Bmc Struct.Biol., 15, 2015
|
|
5W34
 
 | |
5W33
 
 | |
8J2P
 
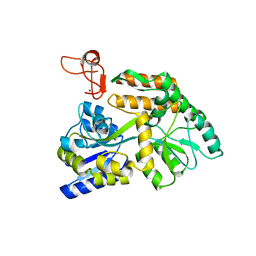 | | Crystal structure of PML B-box2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
8J25
 
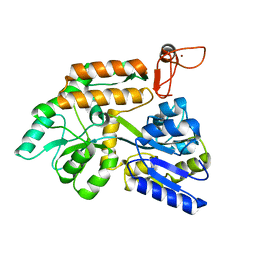 | | Crystal structure of PML B-box2 mutant | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
6S3W
 
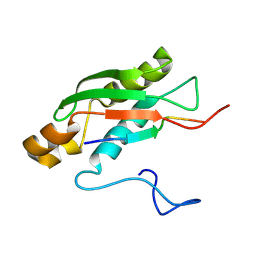 | | Solution NMR Structure of TolAIII Bound to a Peptide Derived from the N-terminus of TolB | | Descriptor: | Cell envelope integrity/translocation protein TolA, TolBp | | Authors: | Kleanthous, C, Redfield, C, Rajasekar, K, Holmes, P. | | Deposit date: | 2019-06-26 | | Release date: | 2020-03-25 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | The lipoprotein Pal stabilises the bacterial outer membrane during constriction by a mobilisation-and-capture mechanism.
Nat Commun, 11, 2020
|
|
7CH6
 
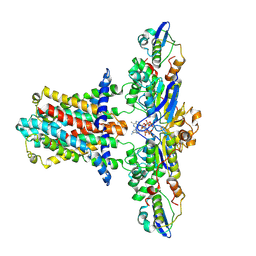 | | Cryo-EM structure of E.coli MlaFEB with AMPPNP | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-08-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH9
 
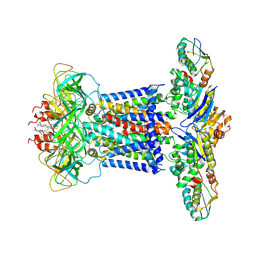 | | Cryo-EM structure of P.aeruginosa MlaFEBD | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, Probable ATP-binding component of ABC transporter, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH8
 
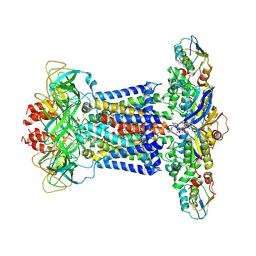 | | Cryo-EM structure of P.aeruginosa MlaFEBD with ADP-V | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CH7
 
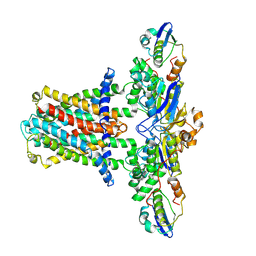 | | Cryo-EM structure of E.coli MlaFEB | | Descriptor: | Lipid asymmetry maintenance ABC transporter permease subunit MlaE, Lipid asymmetry maintenance protein MlaB, Phospholipid ABC transporter ATP-binding protein MlaF | | Authors: | Zhou, C, Shi, H, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7CHA
 
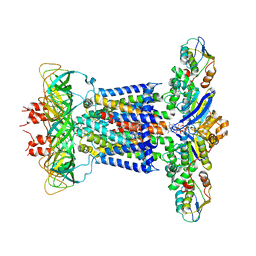 | | Cryo-EM structure of P.aeruginosa MlaFEBD with AMPPNP | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
2LGW
 
 | | Solution Structure of the J Domain of HSJ1a | | Descriptor: | DnaJ homolog subfamily B member 2 | | Authors: | Zhou, C, Gao, X, Cao, C, Hu, H. | | Deposit date: | 2011-08-02 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-terminal helices of heat shock protein 70 are essential for J-domain binding and ATPase activation.
J.Biol.Chem., 287, 2012
|
|
2KLZ
 
 | |
5EAY
 
 | |
5EAW
 
 | | Crystal structure of Dna2 nuclease-helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication ATP-dependent helicase/nuclease DNA2, IRON/SULFUR CLUSTER | | Authors: | Zhou, C, Pourmal, S, Pavletich, N.P. | | Deposit date: | 2015-10-17 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
5EAX
 
 | | Crystal structure of Dna2 in complex with an ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication ATP-dependent helicase/nuclease DNA2, ... | | Authors: | Zhou, C, Pourmal, S, Pavletich, N.P. | | Deposit date: | 2015-10-17 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
5EAN
 
 | | Crystal structure of Dna2 in complex with a 5' overhang DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*CP*TP*CP*TP*GP*CP*CP*AP*AP*GP*AP*GP*GP*A)-3'), ... | | Authors: | Zhou, C, Pourmal, S, Pavletich, N.P. | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
2JWQ
 
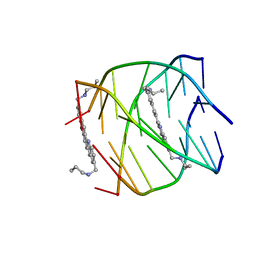 | | G-quadruplex recognition by quinacridines: a SAR, NMR and Biological study | | Descriptor: | DNA (5'-D(*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), N,N'-(dibenzo[b,j][1,7]phenanthroline-2,10-diyldimethanediyl)dipropan-1-amine | | Authors: | Hounsou, C, Guittat, L, Monchaud, D, Jourdan, M, Saettel, N, Mergny, J.L, Teulade-Fichou, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | G-Quadruplex Recognition by Quinacridines: a SAR, NMR, and Biological Study
ChemMedChem, 2, 2007
|
|
7XT2
 
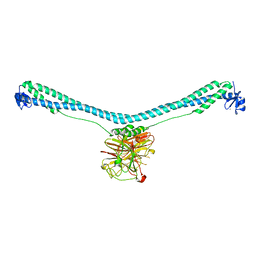 | | Crystal structure of TRIM72 | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Zhou, C, Ma, Y.M, Ding, L. | | Deposit date: | 2022-05-15 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for TRIM72 oligomerization during membrane damage repair.
Nat Commun, 14, 2023
|
|
7XKS
 
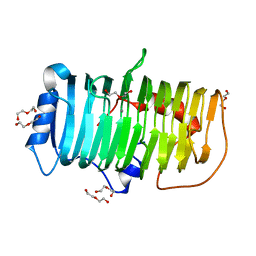 | | Crystal structure of an alkaline pectate lyase from Bacillus clausii | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Pectate lyase | | Authors: | Zhou, C, Zheng, Y.Y, Liu, W.D, Ma, Y. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of an Alkaline Pectate Lyase and Rational Engineering with Improved Thermo-Alkaline Stability for Efficient Ramie Degumming.
Int J Mol Sci, 24, 2022
|
|
2JNH
 
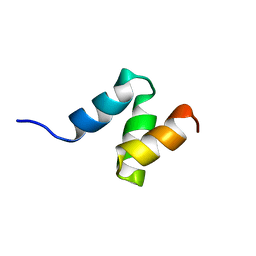 | | Solution Structure of the UBA Domain from Cbl-b | | Descriptor: | E3 ubiquitin-protein ligase CBL-B | | Authors: | Zhou, C, Zhou, Z, Lin, D, Hu, H. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Differential ubiquitin binding of the UBA domains from human c-Cbl and Cbl-b: NMR structural and biochemical insights
Protein Sci., 17, 2008
|
|
2LMG
 
 | |
8IIC
 
 | |
8IIB
 
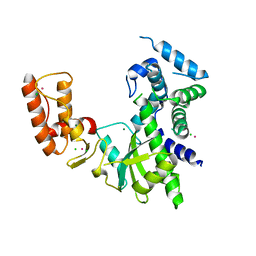 | | Crystal structure of Israeli acute paralysis virus RNA-dependent RNA polymerase delta85 mutant (residues 86-546) | | Descriptor: | CADMIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fang, X, Lu, G, Hou, C, Gong, P. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Unusual substructure conformations observed in crystal structures of a dicistrovirus RNA-dependent RNA polymerase suggest contribution of the N-terminal extension in proper folding.
Virol Sin, 38, 2023
|
|
