7NST
 
 | | ColicinE9 partial translocation complex | | Descriptor: | Colicin-E9, Outer membrane protein F, Tol-Pal system protein TolB | | Authors: | Webby, M.N, Kleanthous, C, Lukoyanova, N, Housden, N.G. | | Deposit date: | 2021-03-08 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Porin threading drives receptor disengagement and establishes active colicin transport through Escherichia coli OmpF.
Embo J., 40, 2021
|
|
3CRQ
 
 | |
5GOR
 
 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 | | Descriptor: | Alkaline Invertase, GLYCEROL, SULFATE ION | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.673 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
5WXZ
 
 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with D-aspartate | | Descriptor: | D-ASPARTIC ACID, McyF | | Authors: | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
3NZ3
 
 | | Crystal structure of the mucin-binding domain of Spr1345 from Streptococcus pneumoniae | | Descriptor: | Putative uncharacterized protein, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Du, Y, He, Y.-X, Zhang, Z.-Y, Yang, Y.-H, Shi, W.-W, Frolet, C, Guilmi, A.M, Vernet, T, Zhou, C.-Z, Chen, Y. | | Deposit date: | 2010-07-15 | | Release date: | 2011-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the mucin-binding domain of Spr1345 from Streptococcus pneumoniae
J.Struct.Biol., 174, 2011
|
|
3TYH
 
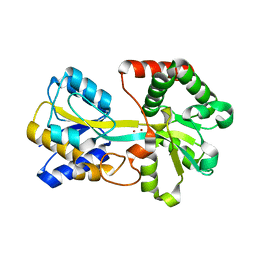 | | Crystal structure of oxo-cupper clusters binding to ferric binding protein from Neisseria gonorrhoeae | | Descriptor: | COPPER (II) ION, FbpA protein | | Authors: | Chen, W.J, Wang, H.F, Zhou, C.J, Ye, D.R, Huang, J, Tan, X.S, Zhong, W.Q. | | Deposit date: | 2011-09-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of oxo-cupper clusters binding to ferric binding protein from Neisseria gonorrhoeae
To be Published
|
|
2P4Q
 
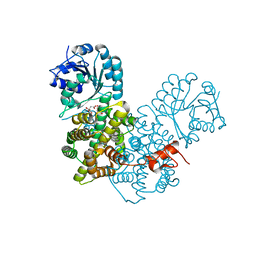 | | Crystal Structure Analysis of Gnd1 in Saccharomyces cerevisiae | | Descriptor: | 6-phosphogluconate dehydrogenase, decarboxylating 1, CITRATE ANION | | Authors: | He, W, Wang, Y, Liu, W, Zhou, C.Z. | | Deposit date: | 2007-03-12 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae 6-phosphogluconate dehydrogenase Gnd1
Bmc Struct.Biol., 7, 2007
|
|
4NT9
 
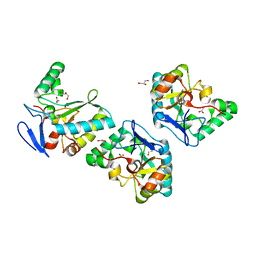 | | Crystal structure of an L,D-carboxypeptidase DacB from Streptococcus pneumonia | | Descriptor: | ACETATE ION, GLYCEROL, Putative uncharacterized protein, ... | | Authors: | Yang, Y.H, Zhang, J, Jiang, Y.L, Zhou, C.Z, Chen, Y. | | Deposit date: | 2013-12-02 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Crystal structure of an L,D-carboxypeptidase DacB from Streptococcus pneumonia
To be Published
|
|
3L4N
 
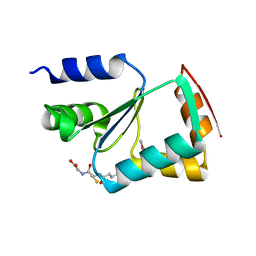 | | Crystal structure of yeast monothiol glutaredoxin Grx6 | | Descriptor: | GLUTATHIONE, Monothiol glutaredoxin-6 | | Authors: | Luo, M, Jiang, Y.-L, Ma, X.-X, He, Y.-X, Tang, Y.-J, Yu, J, Zhang, R.-G, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-04-07 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and biochemical characterization of yeast monothiol glutaredoxin Grx6
J.Mol.Biol., 398, 2010
|
|
8HDS
 
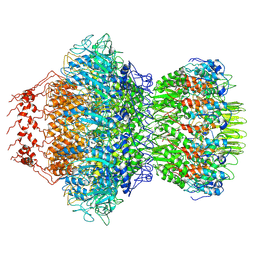 | | Cyanophage Pam3 portal-adaptor | | Descriptor: | Pam3 adaptor protein, Pam3 portal protein | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDT
 
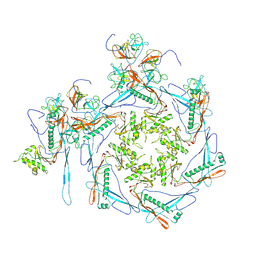 | |
8HDR
 
 | | Cyanophage Pam3 neck | | Descriptor: | Pam3 adaptor protein, Pam3 connector protein, Pam3 sheath protein, ... | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7DMC
 
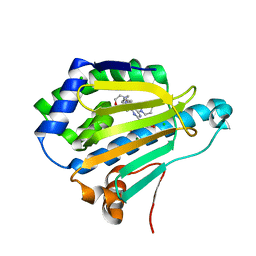 | | Dipyridamole binds to the N-terminal domain of human Hsp90A | | Descriptor: | 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, CHLORIDE ION, Heat shock protein HSP 90-alpha, ... | | Authors: | Shi, L, Zhou, C, Zhong, Y, Gao, J, Zhou, H, Zhang, N. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dipyridamole interacts with the N-terminal domain of HSP90 and antagonizes the function of the chaperone in multiple cancer cell lines.
Biochem Pharmacol, 207, 2022
|
|
7NG9
 
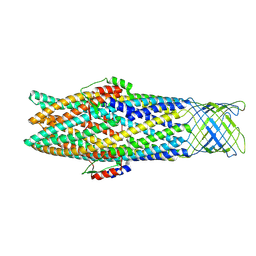 | |
7NNA
 
 | |
4E0H
 
 | | Crystal structure of FAD binding domain of Erv1 from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Mitochondrial FAD-linked sulfhydryl oxidase ERV1 | | Authors: | Guo, P.C, Ma, J.D, Jiang, Y.L, Wang, S.J, Hu, T.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2012-03-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of yeast sulfhydryl oxidase erv1 reveals electron transfer of the disulfide relay system in the mitochondrial intermembrane space
J.Biol.Chem., 287, 2012
|
|
5Z74
 
 | | Crystal structure of alkaline/neutral invertase InvB from Anabaena sp. PCC 7120 complexed with sucrose | | Descriptor: | Alr0819 protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Xie, J, Hu, H.X, Cai, K, Yang, F, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2018-01-27 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and enzymatic analyses of Anabaena heterocyst-specific alkaline invertase InvB.
FEBS Lett., 592, 2018
|
|
8HVH
 
 | | Cryo-EM structure of ABC transporter ABCC3 | | Descriptor: | ATP-binding cassette sub-family C member 3 | | Authors: | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2022-12-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
8HSY
 
 | | Acyl-ACP Synthetase structure | | Descriptor: | Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2022-12-20 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Acyl-ACP Synthetase structure
To Be Published
|
|
7W7H
 
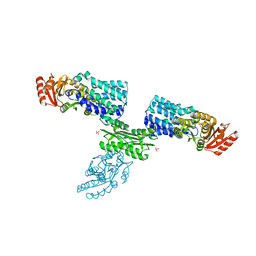 | | S Suis FakA-FakB2 complex structure | | Descriptor: | OLEIC ACID, Predicted kinase related to dihydroxyacetone kinase, SULFATE ION, ... | | Authors: | Shi, Y, Zang, N, Lou, N, Xu, Y, Sun, J, Huang, M, Zhang, H, Lu, H, Zhou, C, Feng, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism for streptococcal fatty acid kinase (Fak) system dedicated to host fatty acid scavenging.
Sci Adv, 8, 2022
|
|
8I22
 
 | | Acyl-ACP synthetase structure bound to pimelic acid monoethyl ester | | Descriptor: | 7-ethoxy-7-oxidanylidene-heptanoic acid, Acyl-acyl carrier protein synthetase | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Acyl-ACP synthetase structure bound to pimelic acid monoethyl ester
To Be Published
|
|
3N5B
 
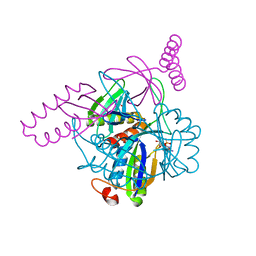 | | The complex of PII and PipX from Anabaena | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Asr0485 protein, Nitrogen regulatory protein P-II, ... | | Authors: | Jiang, Y.-L, Zhao, M.-X, Xu, B.-Y, Chen, Y.-X, Zhang, C.-C, Zhou, C.-Z. | | Deposit date: | 2010-05-24 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the cyanobacterial signal transduction protein PII in complex with PipX.
J.Mol.Biol., 402, 2010
|
|
8I35
 
 | | Acyl-ACP synthetase structure bound to oleic acid | | Descriptor: | Acyl-acyl carrier protein synthetase, OLEIC ACID | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Acyl-ACP synthetase structure bound to oleic acid
To Be Published
|
|
8I51
 
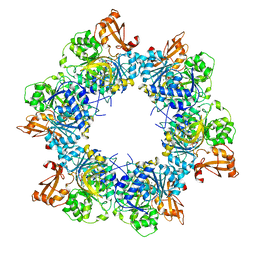 | | Acyl-ACP synthetase structure bound to AMP-MC7 | | Descriptor: | 7-methoxy-7-oxidanylidene-heptanoic acid, ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Acyl-ACP synthetase structure bound to AMP-MC7
To Be Published
|
|
8I3I
 
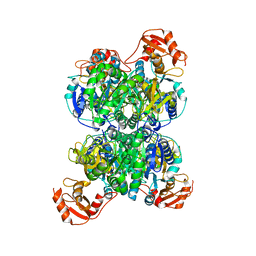 | | Acyl-ACP synthetase structure bound to AMP-PNP in the presence of MgCl2 | | Descriptor: | Acyl-acyl carrier protein synthetase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-17 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Acyl-ACP synthetase structure bound to AMP-PNP in the presence of MgCl2
To Be Published
|
|
