3F3H
 
 | | Crystal structure and anti-tumor activity of LZ-8 from the fungus Ganoderma lucidium | | Descriptor: | Immunomodulatory protein Ling Zhi-8 | | Authors: | Zhou, C.Z, Huang, L, He, Y.X, Bao, R, Sun, F, Liang, C, Liu, L. | | Deposit date: | 2008-10-30 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of LZ-8 from the medicinal fungus Ganoderma lucidium
Proteins, 75, 2009
|
|
5EAY
 
 | |
5EAW
 
 | | Crystal structure of Dna2 nuclease-helicase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication ATP-dependent helicase/nuclease DNA2, IRON/SULFUR CLUSTER | | Authors: | Zhou, C, Pourmal, S, Pavletich, N.P. | | Deposit date: | 2015-10-17 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dna2 nuclease-helicase structure, mechanism and regulation by Rpa.
Elife, 4, 2015
|
|
5XVE
 
 | |
5XU8
 
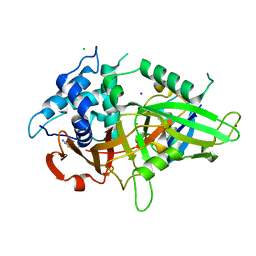 | |
2JNH
 
 | | Solution Structure of the UBA Domain from Cbl-b | | Descriptor: | E3 ubiquitin-protein ligase CBL-B | | Authors: | Zhou, C, Zhou, Z, Lin, D, Hu, H. | | Deposit date: | 2007-01-24 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Differential ubiquitin binding of the UBA domains from human c-Cbl and Cbl-b: NMR structural and biochemical insights
Protein Sci., 17, 2008
|
|
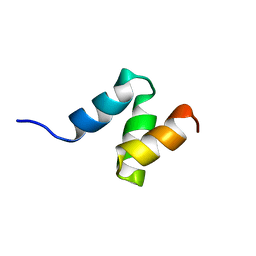
4M0W
 
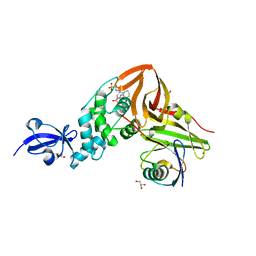 | | Crystal Structure of SARS-CoV papain-like protease C112S mutant in complex with ubiquitin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Replicase polyprotein 1a, ... | | Authors: | Chou, C.-Y, Chen, H.-Y, Lai, H.-Y, Cheng, S.-C, Chou, Y.-W. | | Deposit date: | 2013-08-02 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for catalysis and ubiquitin recognition by the severe acute respiratory syndrome coronavirus papain-like protease
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2LMG
 
 | |
2LGW
 
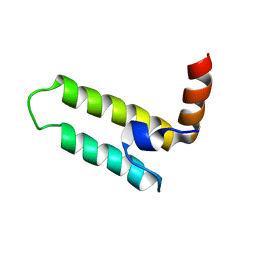 | | Solution Structure of the J Domain of HSJ1a | | Descriptor: | DnaJ homolog subfamily B member 2 | | Authors: | Zhou, C, Gao, X, Cao, C, Hu, H. | | Deposit date: | 2011-08-02 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-terminal helices of heat shock protein 70 are essential for J-domain binding and ATPase activation.
J.Biol.Chem., 287, 2012
|
|
1XE7
 
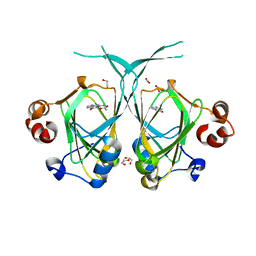 | | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly roll fold | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GUANINE, ... | | Authors: | Zhou, C.-Z, Meyer, P, Quevillon-Cheruel, S, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Leulliot, N, Sorel, I, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2004-09-09 | | Release date: | 2005-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly-roll fold
Protein Sci., 14, 2005
|
|
7XKS
 
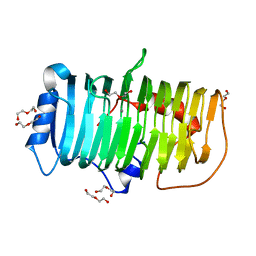 | | Crystal structure of an alkaline pectate lyase from Bacillus clausii | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Pectate lyase | | Authors: | Zhou, C, Zheng, Y.Y, Liu, W.D, Ma, Y. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of an Alkaline Pectate Lyase and Rational Engineering with Improved Thermo-Alkaline Stability for Efficient Ramie Degumming.
Int J Mol Sci, 24, 2022
|
|
7XT2
 
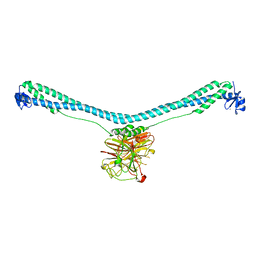 | | Crystal structure of TRIM72 | | Descriptor: | Tripartite motif-containing protein 72, ZINC ION | | Authors: | Zhou, C, Ma, Y.M, Ding, L. | | Deposit date: | 2022-05-15 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for TRIM72 oligomerization during membrane damage repair.
Nat Commun, 14, 2023
|
|
1XE8
 
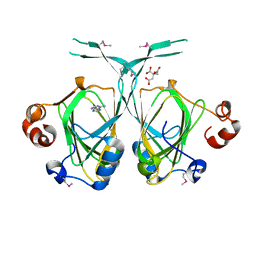 | | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly roll fold. | | Descriptor: | ADENINE, CITRIC ACID, GLYCEROL, ... | | Authors: | Zhou, C.-Z, Meyer, P, Quevillon-Cheruel, S, Li de La Sierra-Gallay, I, Collinet, B, Graille, M, Leulliot, N, Sorel, I, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2004-09-09 | | Release date: | 2005-01-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the YML079w protein from Saccharomyces cerevisiae reveals a new sequence family of the jelly-roll fold
Protein Sci., 14, 2005
|
|
3LCX
 
 | | L-KDO aldolase | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Huang, K.-F, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-12 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
3LCG
 
 | | The D-sialic acid aldolase mutant V251L | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
3LBC
 
 | |
2KLZ
 
 | |
1OCU
 
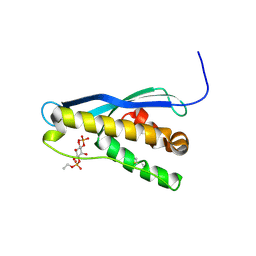 | | Crystal structure of the yeast PX-domain protein Grd19p (sorting nexin 3) complexed to phosphatidylinosytol-3-phosphate. | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, SORTING NEXIN | | Authors: | Zhou, C.Z, Li de La Sierra-Gallay, I, Cheruel, S, Collinet, B, Minard, P, Blondeau, K, Henkes, G, Aufrere, R, Leulliot, N, Graille, M, Sorel, I, Savarin, P, de la Torre, F, Poupon, A, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2003-02-10 | | Release date: | 2003-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate.
J. Biol. Chem., 278, 2003
|
|
1OCS
 
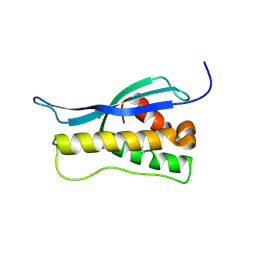 | | Crystal structure of the yeast PX-doamin protein Grd19p (sorting nexin3) complexed to phosphatidylinosytol-3-phosphate. | | Descriptor: | GLYCEROL, SORTING NEXIN GRD19 | | Authors: | Zhou, C.Z, Li De La Sierra-Gallay, I, Cheruel, S, Collinet, B, Minard, P, Blondeau, K, Henkes, G, Aufrere, R, Leulliot, N, Graille, M, Sorel, I, Savarin, P, De La Torre, F, Poupon, A, Janin, J, Van Tilbeurgh, H. | | Deposit date: | 2003-02-10 | | Release date: | 2003-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of the Yeast Phox Homology (Px) Protein Grd19P (Sorting Nexin 3) Complexed to Phosphatidylinositol-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
3LCF
 
 | | The D-sialic acid aldolase mutant V251I | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
5B7C
 
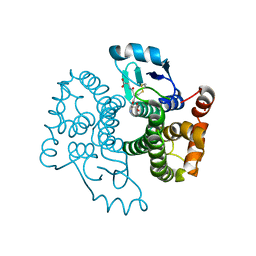 | |
3LCI
 
 | | The D-sialic acid aldolase mutant V251W | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
3LCL
 
 | |
3LCH
 
 | | The D-sialic acid aldolase mutant V251R | | Descriptor: | N-acetylneuraminate lyase, SULFATE ION | | Authors: | Chou, C.-Y, Wang, A.H.-J, Ko, T.-P. | | Deposit date: | 2010-01-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Modulation of substrate specificities of D-sialic acid aldolase through single mutations of Val251
To be Published
|
|
2AN6
 
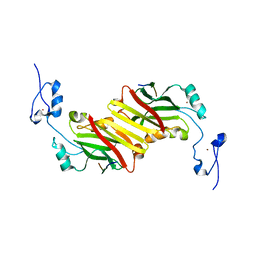 | | Protein-peptide complex | | Descriptor: | Ubiquitin ligase SIAH1A, ZINC ION, peptide from Phyllopod | | Authors: | House, C.M, Hancock, N.C, Moller, A, Cromer, B.A, Fedorov, V, Bowtell, D.D.L, Parker, M.W, Polekhina, G. | | Deposit date: | 2005-08-11 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Elucidation of the substrate binding site of Siah ubiquitin ligase
Structure, 14, 2006
|
|
