3G7A
 
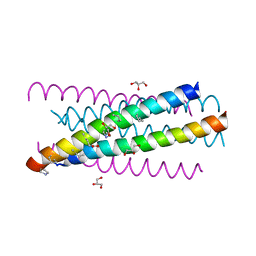 | | HIV gp41 six-helix bundle composed of a chimeric alpha+alpha/beta-peptide analogue of the CHR domain in complex with an NHR domain alpha-peptide | | Descriptor: | ACETYL GROUP, Chimeric alpha+alpha/beta-peptide analogue of the HIV gp41 CHR domain, Envelope glycoprotein gp160, ... | | Authors: | Horne, W.S, Johnson, L.M, Gellman, S.H. | | Deposit date: | 2009-02-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biological mimicry of protein surface recognition by alpha/beta-peptide foldamers
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HF0
 
 | |
3HEZ
 
 | |
6E5J
 
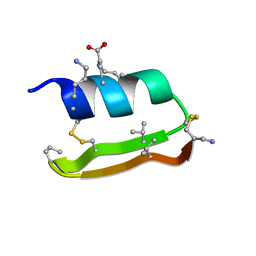 | | Heterogeneous-Backbone Mimics of a Designed Disulfide-Rich Protein: Aib turn, beta3 helix, N-methyl hairpin | | Descriptor: | Designed peptide NC_HEE_D1: Aib turn, beta3 helix, N-methyl hairpin mutant | | Authors: | Cabalteja, C.C, Mihalko, D.S, Horne, W.S. | | Deposit date: | 2018-07-20 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Foldamer Mimics of a Computationally Designed, Disulfide-Rich Miniprotein.
Chembiochem, 20, 2019
|
|
7RAP
 
 | |
8T0G
 
 | |
8T0H
 
 | |
8T0I
 
 | |
6OLN
 
 | |
4A1U
 
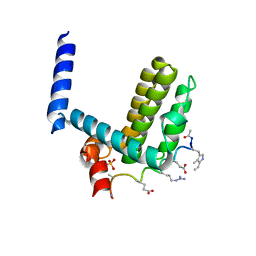 | | Crystal structure of alpha-beta-foldamer 2c in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-FOLDAMER 2C, BCL-2-LIKE PROTEIN 1, CHLORIDE ION, ... | | Authors: | Boersma, M.D, Haase, H.S, Kaufman, K.J, Horne, W.S, Lee, E.F, Clarke, O.B, Smith, B.J, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-09-20 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Evaluation of Diverse Alpha/Beta-Backbone Patterns for Functional Alpha-Helix Mimicry: Analogues of the Bim Bh3 Domain.
J.Am.Chem.Soc., 134, 2012
|
|
4A1W
 
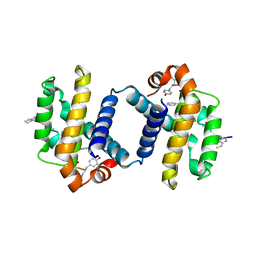 | | Crystal structure of alpha-beta foldamer 4c in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-FOLDAMER 2C, BCL-2-LIKE PROTEIN 1 | | Authors: | Boersma, M.D, Haase, H.S, Kaufman, K.J, Horne, W.S, Lee, E.F, Clarke, O.B, Smith, B.J, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-09-20 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Evaluation of Diverse Alpha/Beta-Backbone Patterns for Functional Alpha-Helix Mimicry: Analogues of the Bim Bh3 Domain.
J.Am.Chem.Soc., 134, 2012
|
|
4TL1
 
 | | GCN4-p1 with mutation to 1-Aminocyclohexanecarboxylic acid at residue 10 | | Descriptor: | General control protein GCN4, ISOPROPYL ALCOHOL | | Authors: | Tavenor, N.A, Silva, K.I, Saxena, S, Horne, W.S. | | Deposit date: | 2014-05-28 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Origins of Structural Flexibility in Protein-Based Supramolecular Polymers Revealed by DEER Spectroscopy.
J.Phys.Chem.B, 118, 2014
|
|
6E5K
 
 | | Heterogeneous-Backbone Mimics of a Designed Disulfide-Rich Protein: Aib turn, Aib helix, N-methyl hairpin | | Descriptor: | Designed peptide NC_HEE_D1: Aib turn, Aib helix, N-methyl hairpin mutant | | Authors: | Cabalteja, C.C, Mihalko, D.S, Horne, W.S. | | Deposit date: | 2018-07-20 | | Release date: | 2018-11-21 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Foldamer Mimics of a Computationally Designed, Disulfide-Rich Miniprotein.
Chembiochem, 20, 2019
|
|
6E5H
 
 | |
6E5I
 
 | |
8G0X
 
 | |
8G0Y
 
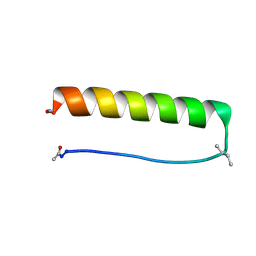 | |
3V3X
 
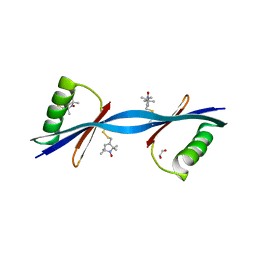 | | Nitroxide Spin Labels in Protein GB1: N8/K28 Double Mutant | | Descriptor: | ACETATE ION, GLYCEROL, Immunoglobulin G-binding protein G, ... | | Authors: | Cunningham, T.F, McGoff, M.S, Sengupta, I, Jaroniec, C.P, Horne, W.S, Saxena, S.K. | | Deposit date: | 2011-12-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of a protein spin-label in a solvent-exposed beta-sheet and comparison with DEER spectroscopy.
Biochemistry, 51, 2012
|
|
5BMI
 
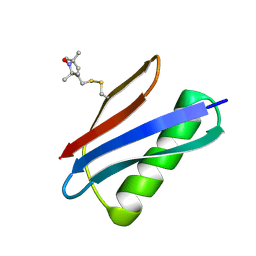 | | Nitroxide Spin Labels in Protein GB1: T44 Mutant, Crystal Form A | | Descriptor: | Immunoglobulin G-binding protein G, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Cunningham, T.C, Horne, W.S, Saxena, S. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rotameric preferences of a protein spin label at edge-strand beta-sheet sites.
Protein Sci., 25, 2016
|
|
5BMH
 
 | | Nitroxide Spin Labels in Protein GB1: T44 Mutant, Crystal Form B | | Descriptor: | Immunoglobulin G-binding protein G, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Cunningham, T.C, Horne, W.S, Saxena, S. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rotameric preferences of a protein spin label at edge-strand beta-sheet sites.
Protein Sci., 25, 2016
|
|
5BMG
 
 | | Nitroxide Spin Labels in Protein GB1: E15 Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Immunoglobulin G-binding protein G, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Cunningham, T.C, Horne, W.S, Saxena, S. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rotameric preferences of a protein spin label at edge-strand beta-sheet sites.
Protein Sci., 25, 2016
|
|
8ERZ
 
 | |
8ERY
 
 | |
8ES2
 
 | |
8ES3
 
 | |
