3SNF
 
 | | Onconase, atomic resolution crystal structure | | Descriptor: | ACETATE ION, Protein P-30, SULFATE ION | | Authors: | Holloway, D.E, Singh, U.P, Shogen, K, Acharya, K.R. | | Deposit date: | 2011-06-29 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of Onconase at 1.1 angstrom resolution--insights into substrate binding and collective motion.
Febs J., 278, 2011
|
|
1GV7
 
 | | ARH-I, an angiogenin/RNase A chimera | | Descriptor: | ANGIOGENIN, CITRIC ACID | | Authors: | Holloway, D.E, Shapiro, R, Hares, M.C, Leonidas, D.D, Acharya, K.R. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Guest-Host Crosstalk in an Angiogenin-RNase A Chimeric Protein
Biochemistry, 41, 2002
|
|
1UN5
 
 | | ARH-II, AN ANGIOGENIN/RNASE A CHIMERA | | Descriptor: | ANGIOGENIN, CITRIC ACID | | Authors: | Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2003-09-04 | | Release date: | 2004-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Studies on Structural Features that Determine the Enzymatic Specificity and Potency of Human Angiogenin: Thr44, Thr80 and Residues 38-41
Biochemistry, 43, 2004
|
|
1UN3
 
 | |
1UN4
 
 | |
2BWL
 
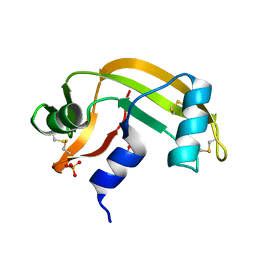 | | Murine angiogenin, phosphate complex | | Descriptor: | ANGIOGENIN, PHOSPHATE ION | | Authors: | Holloway, D.E, Chavali, G.B, Hares, M.C, Subramanian, V, Acharya, K.R. | | Deposit date: | 2005-07-15 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of Murine Angiogenin: Features of the Substrate- and Cell-Binding Regions and Prospects for Inhibitor-Binding Studies.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2BWK
 
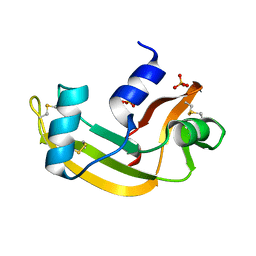 | | Murine angiogenin, sulphate complex | | Descriptor: | ANGIOGENIN, SULFATE ION | | Authors: | Holloway, D.E, Chavali, G.B, Hares, M.C, Subramanian, V, Acharya, K.R. | | Deposit date: | 2005-07-15 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Murine Angiogenin: Features of the Substrate- and Cell-Binding Regions and Prospects for Inhibitor-Binding Studies.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3ZBV
 
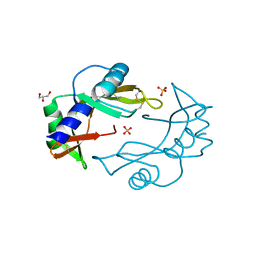 | | Crystal Structure of murine Angiogenin-2 | | Descriptor: | ANGIOGENIN-2, GLYCEROL, SULFATE ION, ... | | Authors: | Iyer, S, Holloway, D.E, Acharya, K.R. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of Murine Angiogenin-2 and -3 - Probing 'Structure - Function' Relationships Amongst Angiogenin Homologues.
FEBS J., 280, 2013
|
|
3ZBW
 
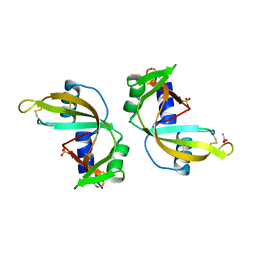 | | Crystal Structure of murine Angiogenin-3 | | Descriptor: | ACETIC ACID, ANGIOGENIN-3, SULFATE ION, ... | | Authors: | Iyer, S, Holloway, D.E, Acharya, K.R. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structures of Murine Angiogenin-2 and -3 - Probing 'Structure - Function' Relationships Amongst Angiogenin Homologues.
FEBS J., 280, 2013
|
|
2J4T
 
 | | Biological and Structural Features of Murine Angiogenin-4, an Angiogenic Protein | | Descriptor: | ANGIOGENIN-4 | | Authors: | Crabtree, B, Holloway, D.E, Baker, M.D, Acharya, K.R, Subramanian, V. | | Deposit date: | 2006-09-05 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biological and Structural Features of Murine Angiogenin-4, an Angiogenic Protein
Biochemistry, 46, 2007
|
|
1GQV
 
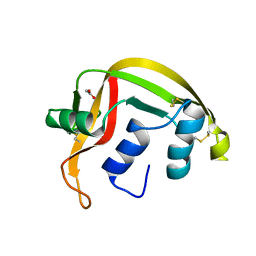 | | Atomic Resolution (0.98A) Structure of Eosinophil-Derived Neurotoxin | | Descriptor: | ACETATE ION, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Swaminathan, G.J, Holloway, D.E, Veluraja, K, Acharya, K.R. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution (0.98 A) Structure of Eosinophil-Derived Neurotoxin
Biochemistry, 41, 2002
|
|
1UZI
 
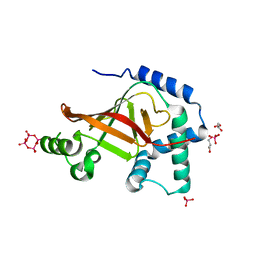 | | C3 EXOENZYME FROM CLOSTRIDIUM BOTULINUM, TETRAGONAL FORM | | Descriptor: | CYCLO-TETRAMETAVANADATE, GLYCEROL, MONO-ADP-RIBOSYLTRANSFERASE C3, ... | | Authors: | Evans, H.R, Holloway, D.E, Sutton, J.M, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | C3 Exoenzyme from Clostridium Botulinum: Structure of a Tetragonal Crystal Form and a Reassessment of Nad-Induced Flexure
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OJQ
 
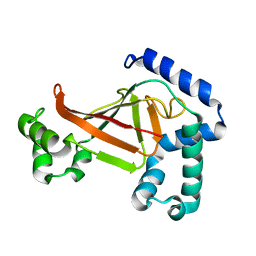 | | The crystal structure of C3stau2 from S. aureus | | Descriptor: | ADP-RIBOSYLTRANSFERASE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-15 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
1OJZ
 
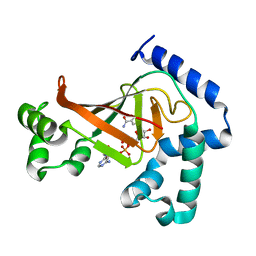 | | The crystal structure of C3stau2 from S. aureus with NAD | | Descriptor: | ADP-RIBOSYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Evans, H.R, Sutton, J.M, Holloway, D.E, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2003-07-16 | | Release date: | 2003-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Crystal Structure of C3Stau2 from Staphylococcus Aureus and its Complex with Nad
J.Biol.Chem., 278, 2003
|
|
1O7Z
 
 | |
1O7Y
 
 | | Crystal structure of IP-10 M-form | | Descriptor: | SMALL INDUCIBLE CYTOKINE B10, SULFATE ION | | Authors: | Swaminathan, G.J, Holloway, D.E, Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 2002-11-20 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Oligomeric Forms of the Ip-10/Cxcl10 Chemokine
Structure, 11, 2003
|
|
1O80
 
 | |
2BZZ
 
 | | Crystal Structures of Eosinophil-derived Neurotoxin in Complex with the Inhibitors 5'-ATP, Ap3A, Ap4A and Ap5A | | Descriptor: | ACETIC ACID, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, NONSECRETORY RIBONUCLEASE | | Authors: | Baker, M.D, Holloway, D.E, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2005-08-24 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystal Structures of Eosinophil-Derived Neurotoxin (Edn) in Complex with the Inhibitors 5'- ATP, Ap(3)A, Ap(4)A, and Ap(5)A.
Biochemistry, 45, 2006
|
|
2C02
 
 | | Crystal Structures of Eosinophil-derived Neurotoxin in Complex with the Inhibitors 5'-ATP, Ap3A, Ap4A and Ap5A | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, NONSECRETORY RIBONUCLEASE | | Authors: | Baker, M.D, Holloway, D.E, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2005-08-24 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Eosinophil-Derived Neurotoxin (Edn) in Complex with the Inhibitors 5'- ATP, Ap(3)A, Ap(4)A, and Ap(5)A.
Biochemistry, 45, 2006
|
|
2C05
 
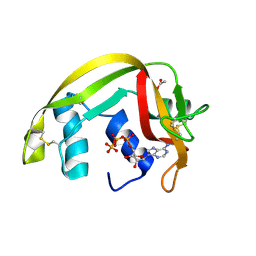 | | Crystal Structures of Eosinophil-derived Neurotoxin in Complex with the Inhibitors 5'-ATP, Ap3A, Ap4A and Ap5A | | Descriptor: | ACETIC ACID, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, NONSECRETORY RIBONUCLEASE | | Authors: | Baker, M.D, Holloway, D.E, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2005-08-25 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structures of Eosinophil-Derived Neurotoxin (Edn) in Complex with the Inhibitors 5'- ATP, Ap(3)A, Ap(4)A, and Ap(5)A.
Biochemistry, 45, 2006
|
|
2BEX
 
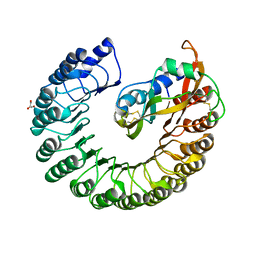 | | Crystal structure of Placental Ribonuclease Inhibitor in complex with Human Eosinophil Derived Neurotoxin at 2A resolution | | Descriptor: | ALPHA-KETOMALONIC ACID, GLYCEROL, NONSECRETORY RIBONUCLEASE, ... | | Authors: | Iyer, S, Holloway, D.E, Kumar, K, Shapiro, R, Acharya, K.R. | | Deposit date: | 2004-12-01 | | Release date: | 2005-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular Recognition of Human Eosinophil-Derived Neurotoxin (Rnase 2) by Placental Ribonuclease Inhibitor
J.Mol.Biol., 347, 2005
|
|
2C01
 
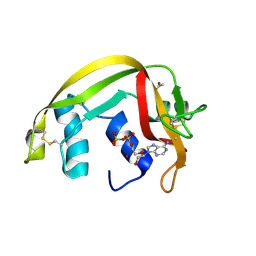 | | Crystal Structures of Eosinophil-derived Neurotoxin in Complex with the Inhibitors 5'-ATP, Ap3A, Ap4A and Ap5A | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, NONSECRETORY RIBONUCLEASE | | Authors: | Baker, M.D, Holloway, D.E, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2005-08-24 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal Structures of Eosinophil-Derived Neurotoxin (Edn) in Complex with the Inhibitors 5'- ATP, Ap(3)A, Ap(4)A, and Ap(5)A.
Biochemistry, 45, 2006
|
|
2VQ8
 
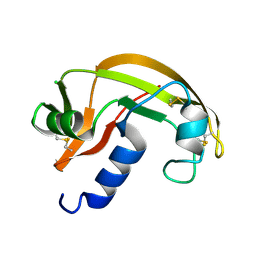 | | RNASE ZF-1A | | Descriptor: | CHLORIDE ION, RNASE ZF-1A | | Authors: | Kazakou, K, Holloway, D.E, Prior, S.H, Subramanian, V, Acharya, K.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Ribonuclease A Homologues of the Zebrafish: Polymorphism, Crystal Structures of Two Representatives and Their Evolutionary Implications.
J.Mol.Biol., 380, 2008
|
|
2VQ9
 
 | | RNASE ZF-3E | | Descriptor: | CHLORIDE ION, RNASE 1 | | Authors: | Kazakou, K, Holloway, D.E, Prior, S.H, Subramanian, V, Acharya, K.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ribonuclease A Homologues of the Zebrafish: Polymorphism, Crystal Structures of Two Representatives and Their Evolutionary Implications
J.Mol.Biol., 380, 2008
|
|
2W5K
 
 | | RNASE A-NADPH COMPLEX | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RIBONUCLEASE PANCREATIC | | Authors: | Chavali, G.B, Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influence of Naturally-Occurring 5'-Pyrophosphate-Linked Substituents on the Binding of Adenylic Inhibitors to Ribonuclease A: An X-Ray Crystallographic Study.
Biopolymers, 91, 2009
|
|
